Installation
The natverse runs on top of R. If you have never installed R see the First time install and Step by Step illustrated guide at the bottom of this page.
Basic Install
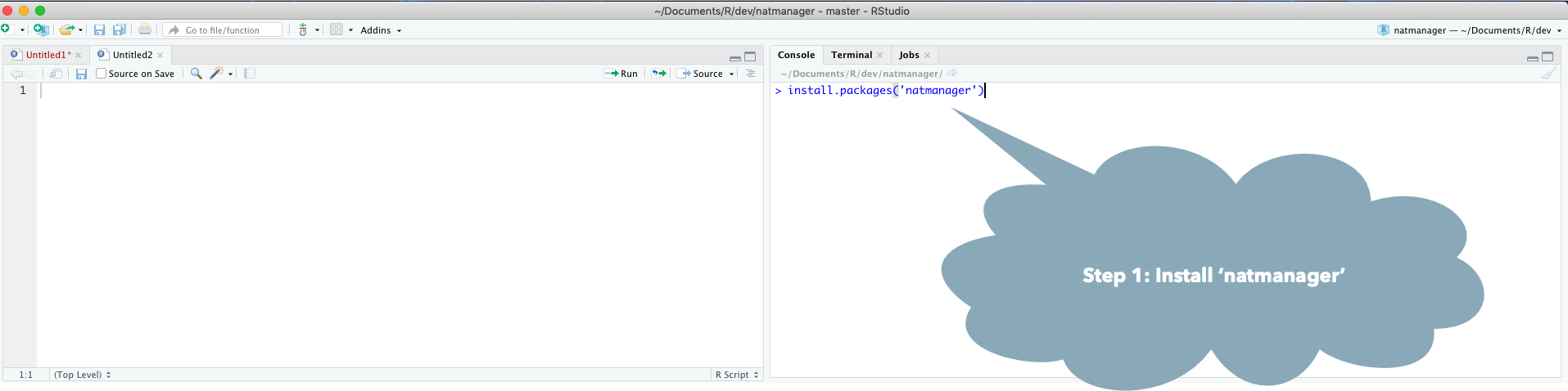
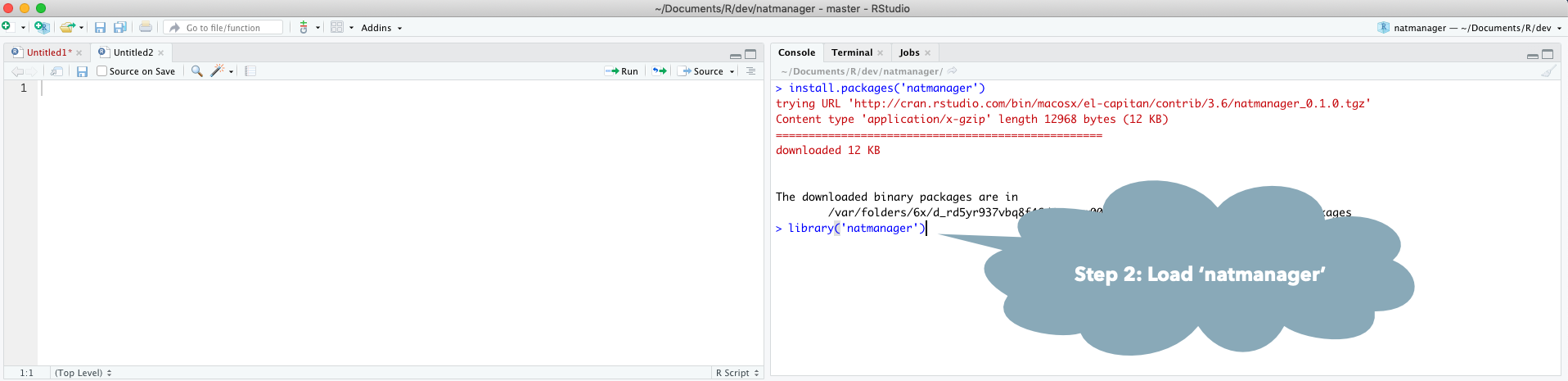
Install core natverse packages to get started by doing
install.packages("natmanager")
natmanager::install('core')
Full Install
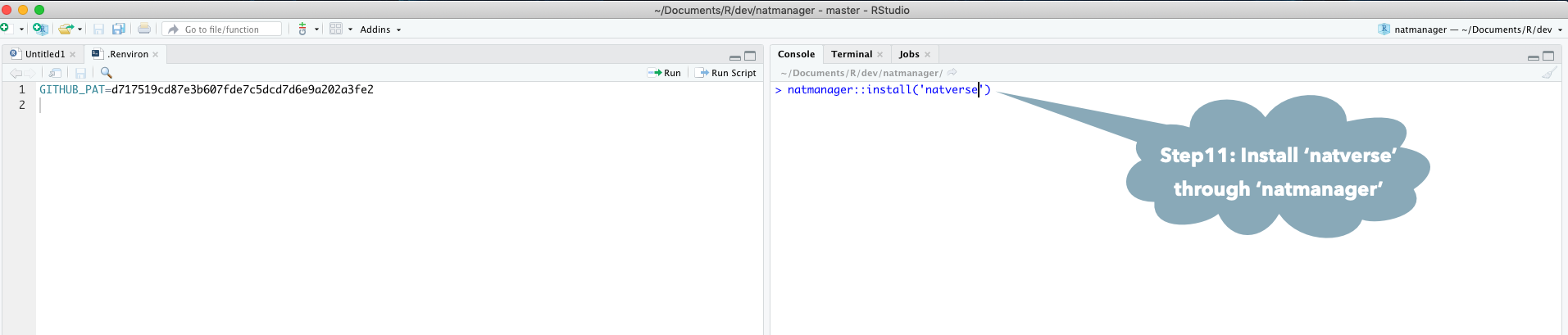
Install all the packages in the natverse by running:
install.packages("natmanager")
natmanager::install('natverse')
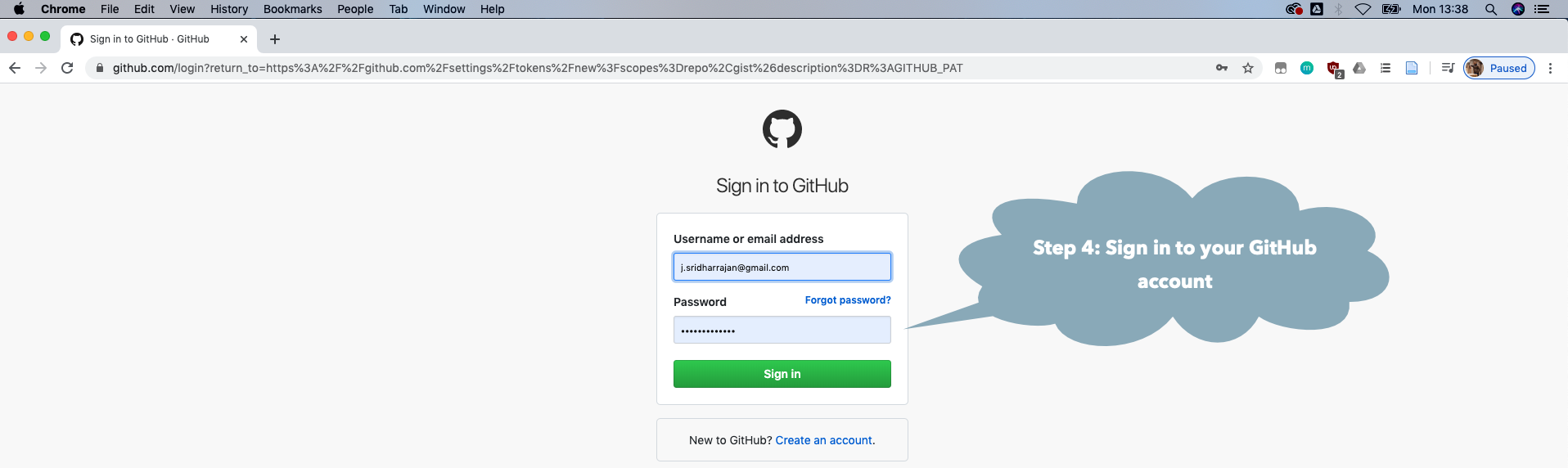
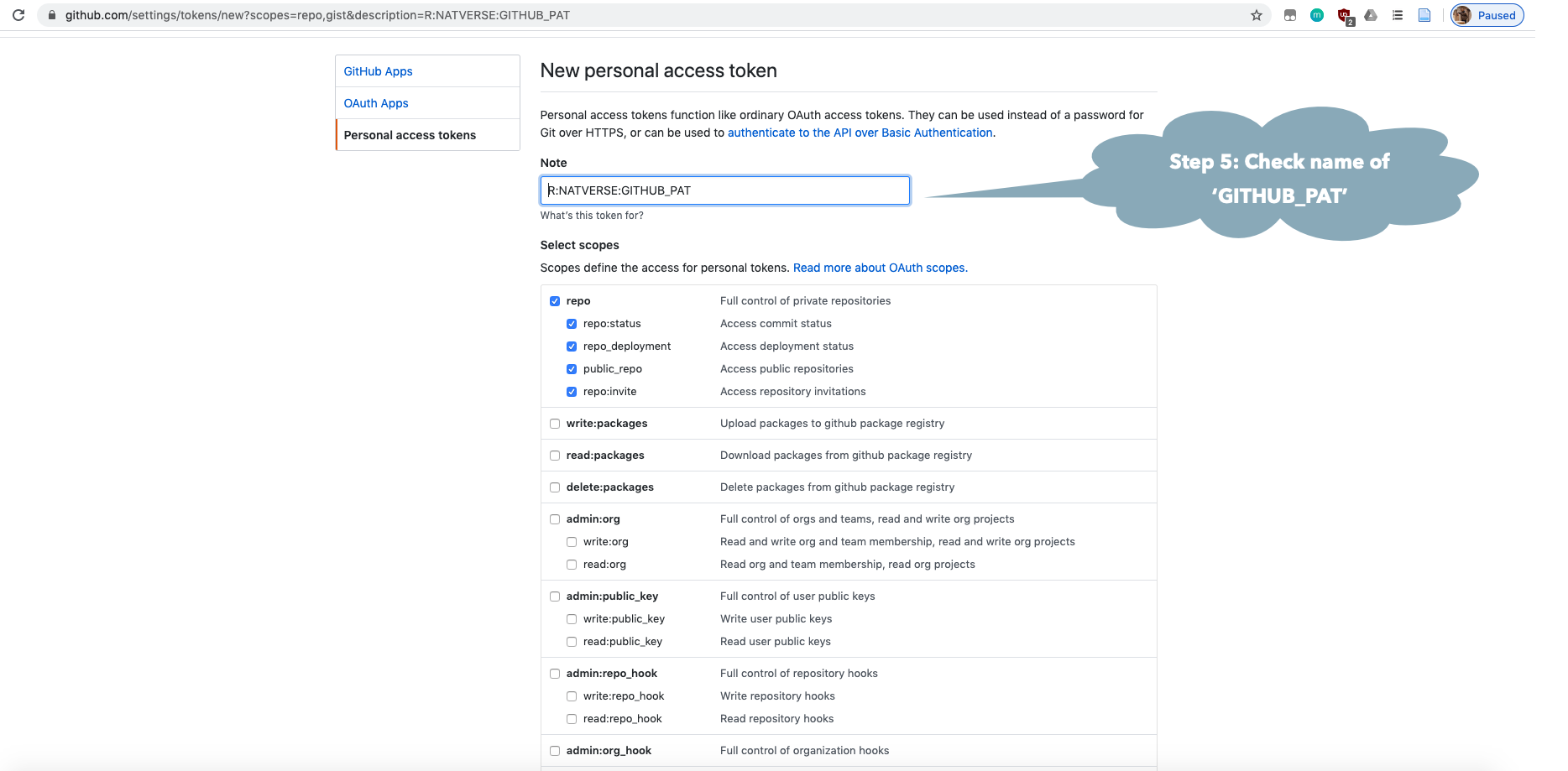
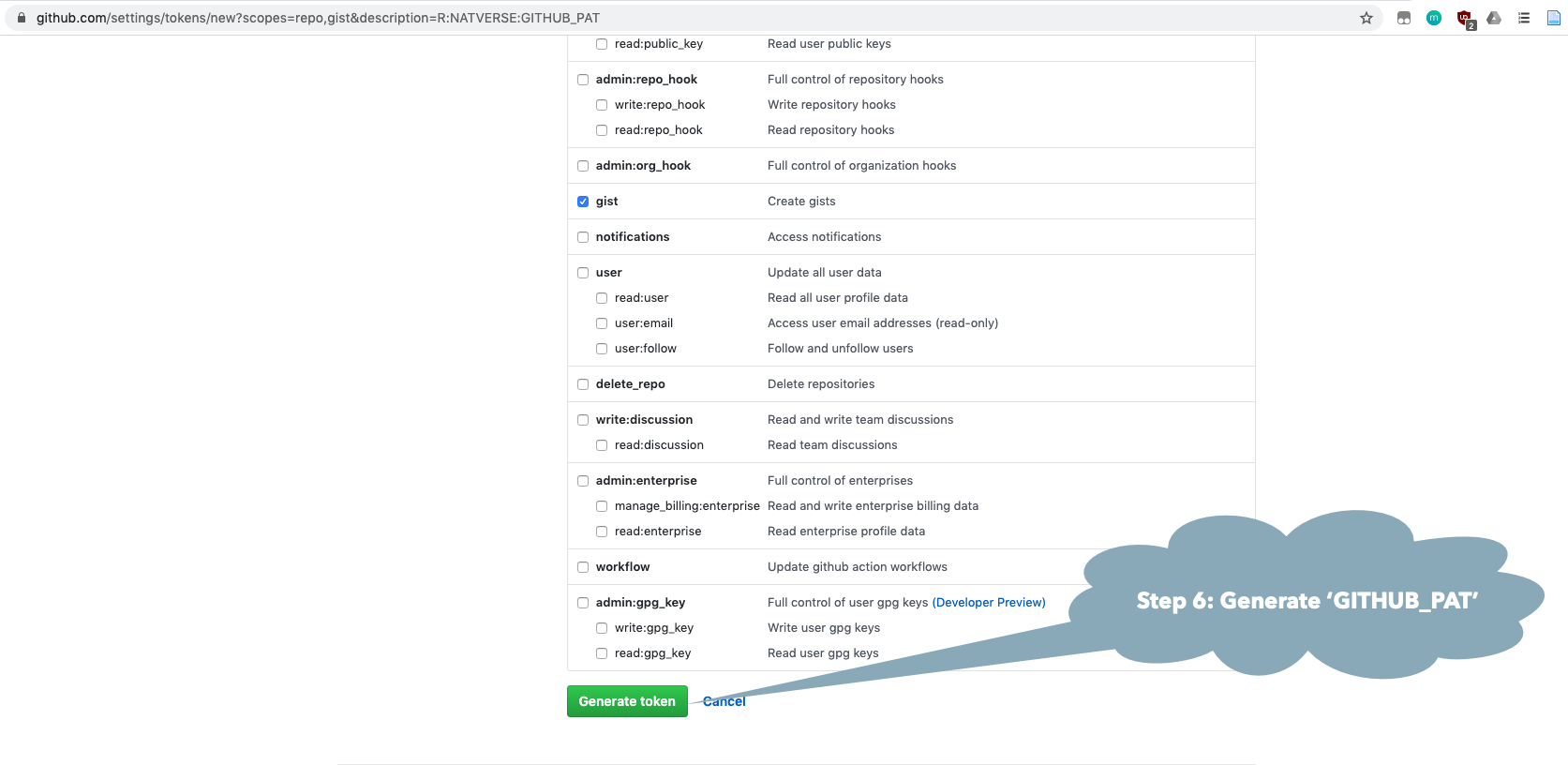
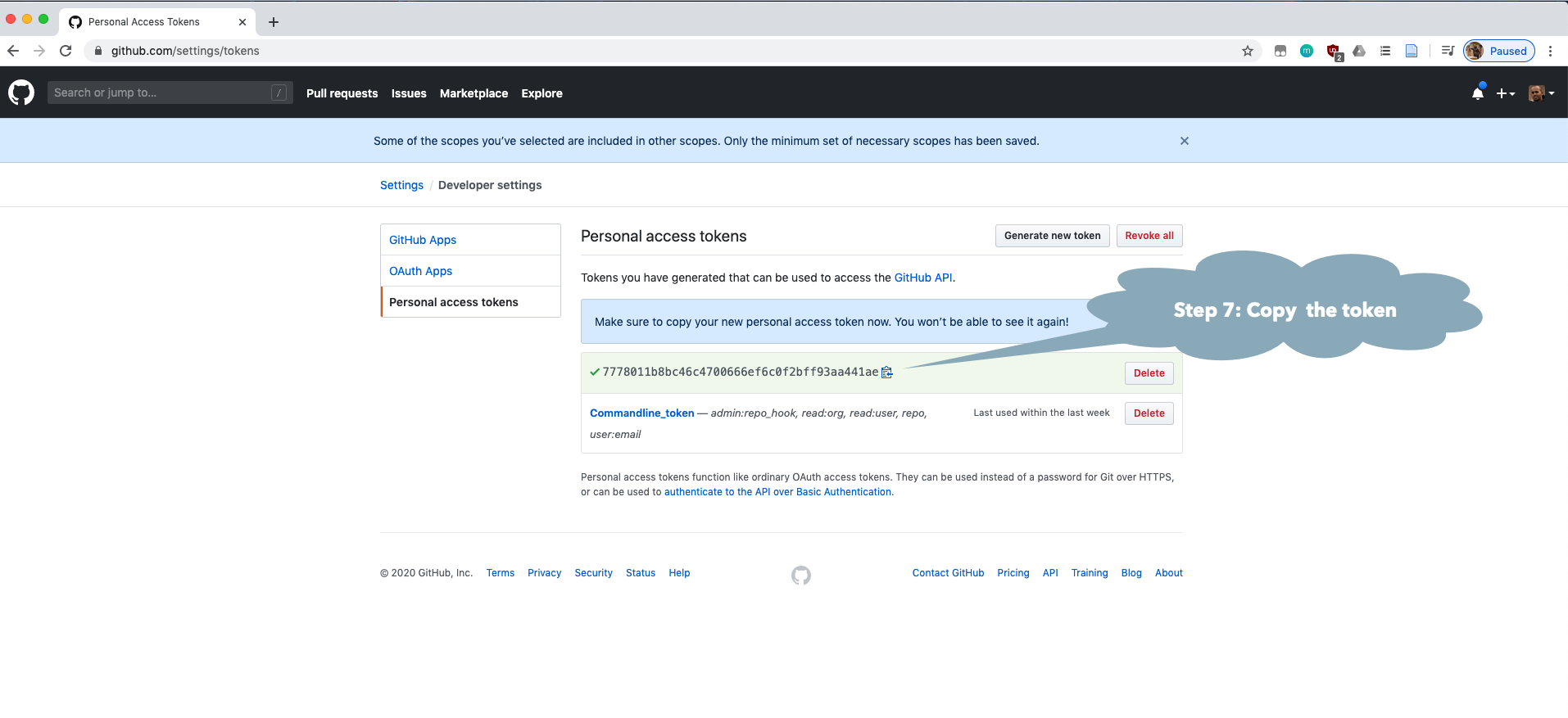
You may need to make a GitHub account and personal access token (GITHUB_PAT) if you have not done this previously. See the GitHub Rate limiting and Step by Step Installation Guide sections below.
Run
To load the core natverse and make it available in your current R session:
library(natverse)
You can check everything is working with some simple plots:
# plot some test data (?kcs20 for details)
# Drosophila Kenyon cells processed from raw data at http://flycircuit.tw
head(kcs20)
open3d()
plot3d(kcs20, col=type)
# get help
?nat
To find our more, explore our resources for learning the natverse.
Update
To update all natverse packages (and their dependencies):
library(natverse)
# see what needs updating
natverse_update()
# actually update
natverse_update(update = TRUE)
# ... or if you don't want to answer any yes/no questions
natverse_update(update = TRUE, upgrade='always')
Troubleshooting
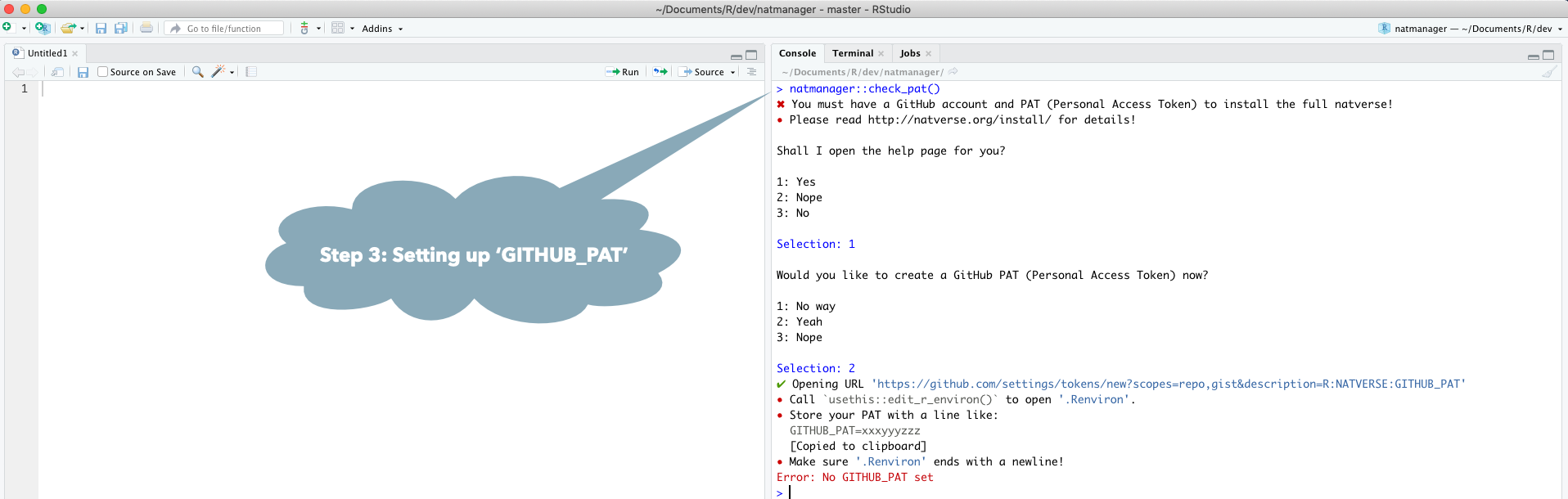
GitHub Rate limiting
Unfortunately installing the full natverse suite makes many calls to the GitHub
API, which is rate limited unless you have a GitHub account and present a
GITHUB_PAT authentication token. The natmanager package will attempt to
use a built-in PAT if you do not have one. However, if multiple users are
installing the natverse simultaneously, you may get errors like
Using github PAT from envvar GITHUB_PAT
Error: Failed to install 'unknown package' from GitHub:
HTTP error 401.
Bad credentials
Rate limit remaining: 56/60
Rate limit reset at: 2020-02-27 19:21:26 UTC
To avoid these errors, do
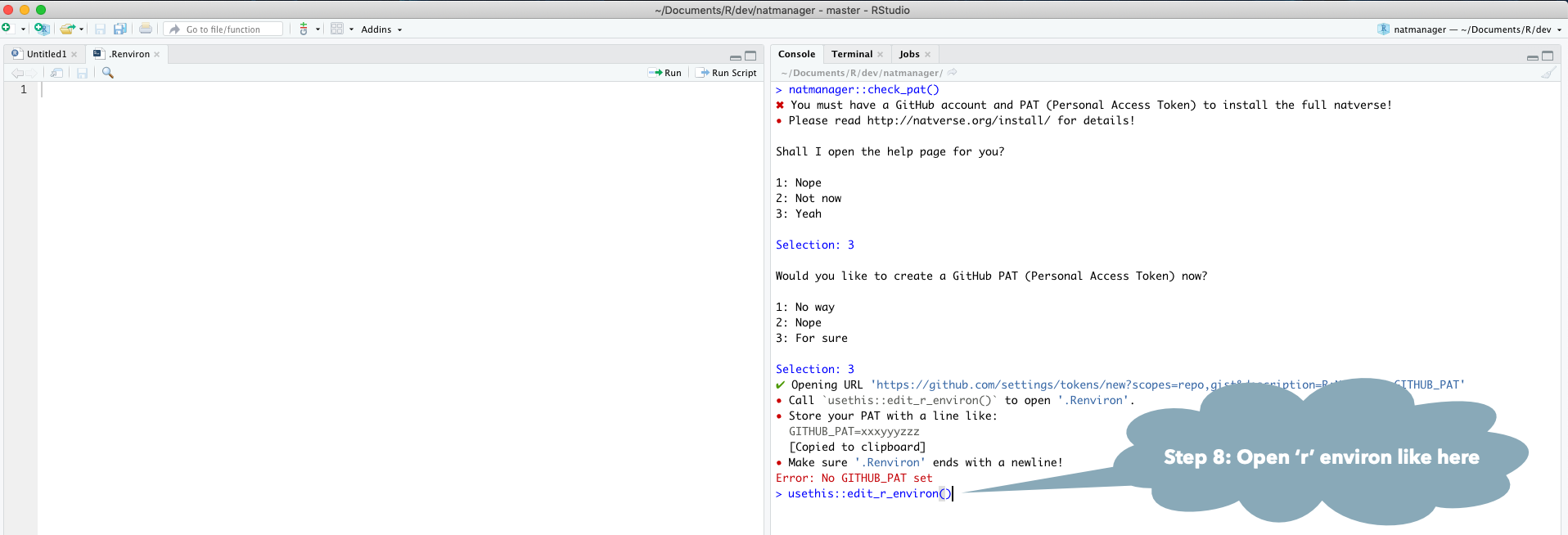
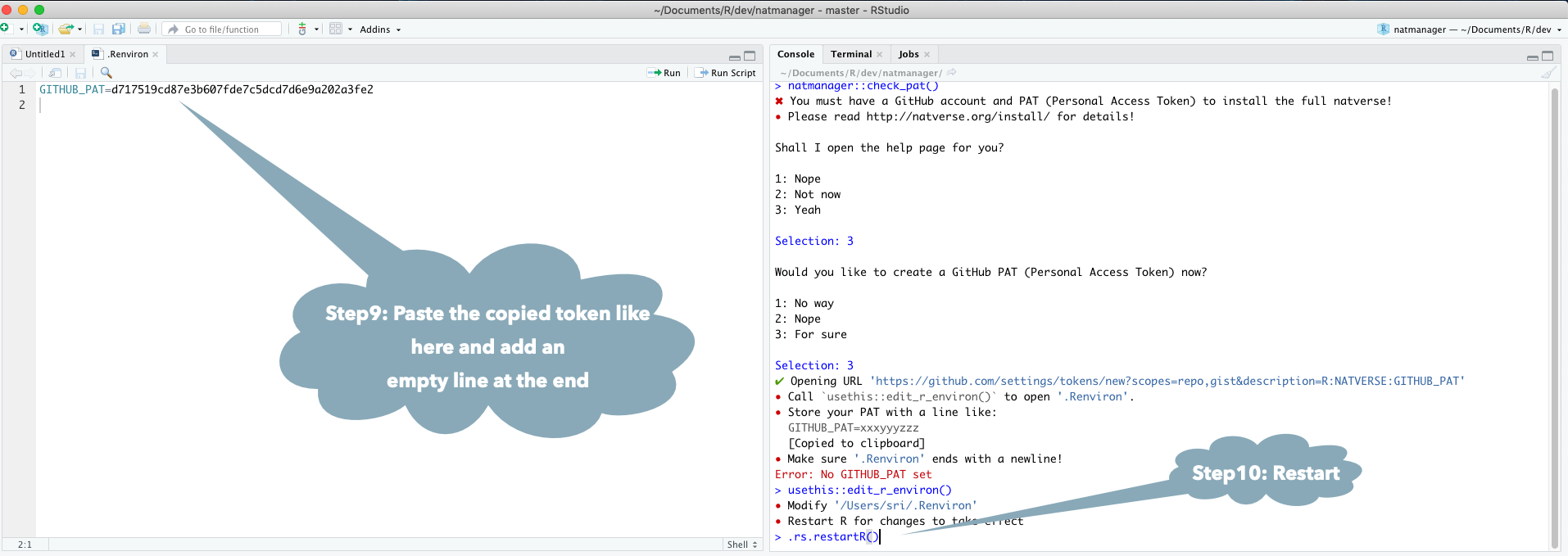
natmanager::check_pat()
and this will guide you through the process of obtaining your own GITHUB_PAT
and telling R about it. See also the illustrated guide below.
Other install issues
If you have any installation issues, some general tips:
- Start a clean R session
- Start a clean R session if you had to update natmanager
- make sure that you have recent R (>=3.6.0 recommended, R>=3.3.0 at a minimum)
- try updating installed packages (
update.packages(ask=F)will update everything)
Still no luck?
- most install issues will be general R issues rather than anything specific to the natverse, so Google is your friend.
- Search the nat-user google group.
- … or use it to ask for help.
First time install - Prerequisites
To install this package you need to have R and RStudio installed. Detailed
instructions for each operating system are given below. You will also need a
valid GitHub account to install the full suite of natverse packages. You will be
prompted to do this by the natmanager package, but we recommend following the
illustrated Step by Step Installation Guide.
Mac OS X
Install
Rfrom here: r-installationInstall
RStudio, from here: RStudio-installationStart
RStudiofrom LaunchpadTo use the rgl package for 3D visualisation you may also need to install the Xquartz X11 system, which is also available by following links from the CRAN Mac OS X page.
Windows
Install
Rfrom here: r-installationInstall
RStudio, for Windows from here: RStudio-installationStart
RStudiofrom Programs menu
Linux (Ubuntu)
Install
R, in terminal type below:sudo apt-get install libopenblas-base r-baseInstall
RStudio, in terminal type below:sudo apt-get install gdebi cd ~/Downloads wget https://download1.rstudio.org/rstudio-xenial-1.1.419-amd64.deb sudo gdebi rstudio-xenial-1.1.419-amd64.debStart
RStudio, in terminal type below:rstudio
Step by Step Installation Guide
By: Sridhar Jagannathan & Gregory Jefferis