Get connectivity adjacency matrix between set of neurons
Source:R/catmaid_skeleton.R
catmaid_adjacency_matrix.RdGet connectivity adjacency matrix between set of neurons
catmaid_adjacency_matrix(
inputskids,
outputskids = inputskids,
pid = 1,
conn = NULL,
...
)Arguments
- inputskids, outputskids
Input (source) and output (target) skids in any form understandable to
catmaid_skids.- pid
Project id (default 1)
- conn
A
catmaid_connectionobjection returned bycatmaid_login. IfNULL(the default) a new connection object will be generated using the values of the catmaid.* package options as described in the help forcatmaid_login.- ...
Additional arguments passed to the
catmaid_fetchfunction
Value
A matrix, with inputs (sources) as rows and outputs (targets) as columns.
See also
Other connectors:
catmaid_get_connector_table(),
catmaid_get_connectors_between(),
catmaid_get_connectors(),
catmaid_query_connected(),
connectors()
Examples
# \donttest{
conn=vfbcatmaid('fafb')
da1adj=catmaid_adjacency_matrix("name:DA1", conn=conn)
# note that we translate skids to neuron names (longer but more informative)
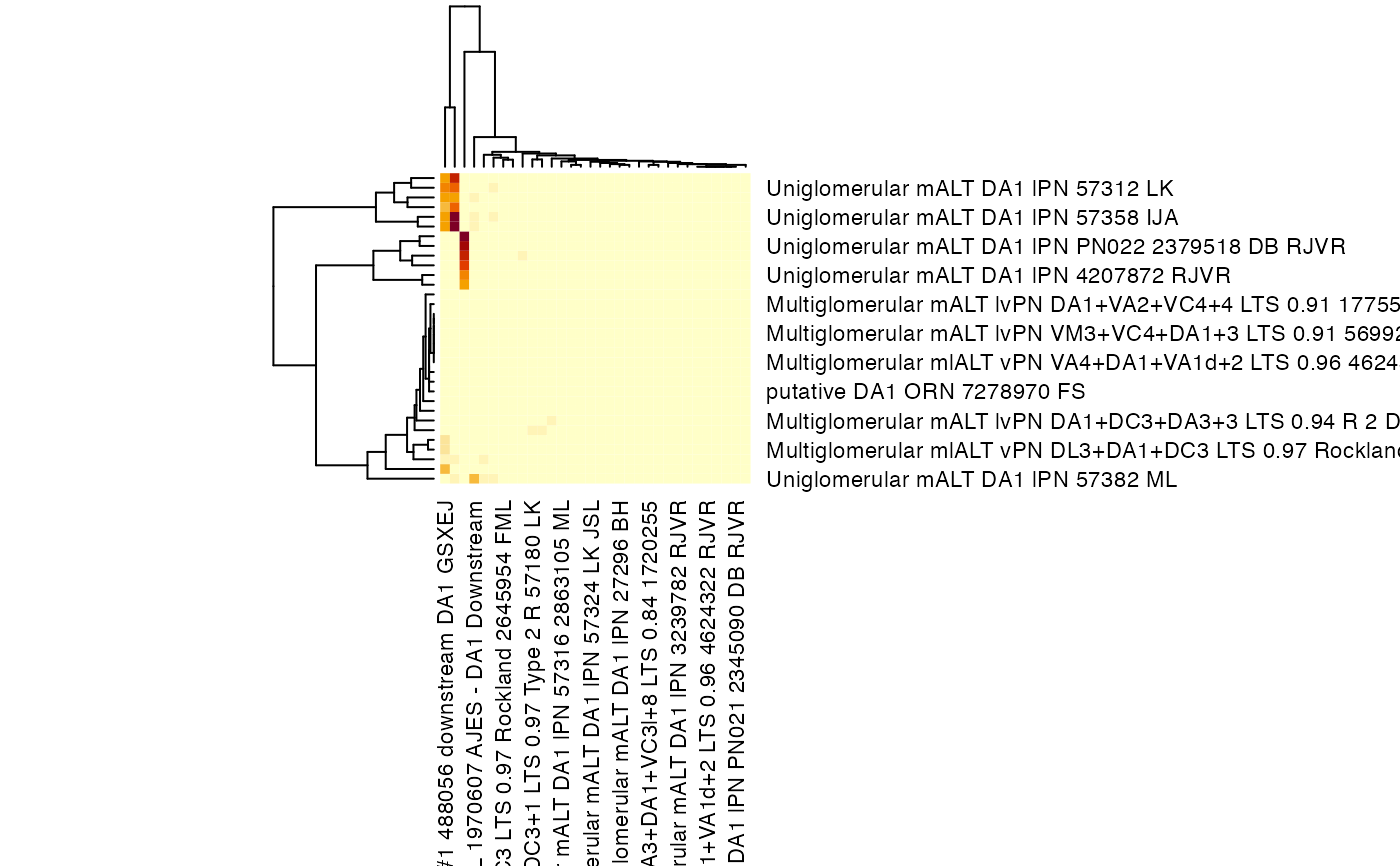
heatmap(
da1adj,
scale = 'none',
labCol = catmaid_get_neuronnames(colnames(da1adj)),
labRow = catmaid_get_neuronnames(rownames(da1adj)),
margins = c(12,12)
)
 # }
# }