Methods to identify and plot groups of neurons cut from an hclust object
Source: R/clustering.r
plot3d.hclust.Rdplot3d.hclust uses plot3d to plot neurons from
each group, cut from the hclust object, by colour.
Usage
# S3 method for class 'hclust'
plot3d(
x,
k = NULL,
h = NULL,
groups = NULL,
col = rainbow,
colour.selected = FALSE,
...
)Arguments
- x
- k
number of clusters to cut from
hclustobject.- h
height to cut
hclustobject.- groups
numeric vector of groups to plot.
- col
colours for groups (directly specified or a function).
- colour.selected
When set to
TRUEthe colour palette only applies to the displayed cluster groups (defaultFALSE).- ...
additional arguments for
plot3d
Value
A list of rgl IDs for plotted objects (see
plot3d).
Examples
# 20 Kenyon cells
data(kcs20, package='nat')
# calculate mean, normalised NBLAST scores
kcs20.aba=nblast_allbyall(kcs20)
kcs20.hc=nhclust(scoremat = kcs20.aba)
#> The "ward" method has been renamed to "ward.D"; note new "ward.D2"
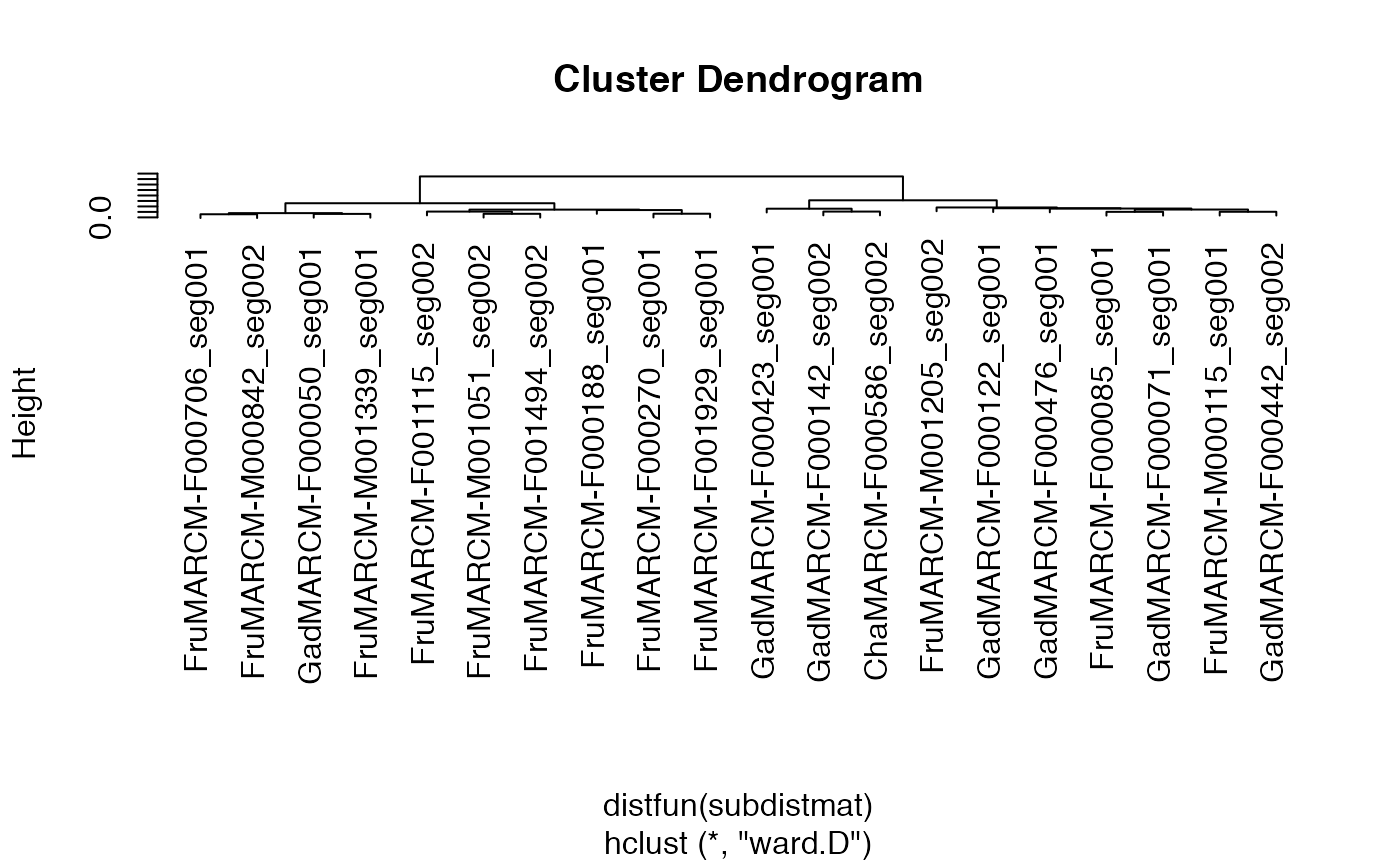
# plot the resultant dendrogram
plot(kcs20.hc)
 # now plot the neurons in 3D coloured by cluster group
# note that specifying db explicitly could be avoided by use of the
# \code{nat.default.neuronlist} option.
plot3d(kcs20.hc, k=3, db=kcs20)
# only plot first two groups
# (will plot in same colours as when all groups are plotted)
plot3d(kcs20.hc, k=3, db=kcs20, groups=1:2)
# only plot first two groups
# (will be coloured with a two-tone palette)
plot3d(kcs20.hc, k=3, db=kcs20, groups=1:2, colour.selected=TRUE)
# now plot the neurons in 3D coloured by cluster group
# note that specifying db explicitly could be avoided by use of the
# \code{nat.default.neuronlist} option.
plot3d(kcs20.hc, k=3, db=kcs20)
# only plot first two groups
# (will plot in same colours as when all groups are plotted)
plot3d(kcs20.hc, k=3, db=kcs20, groups=1:2)
# only plot first two groups
# (will be coloured with a two-tone palette)
plot3d(kcs20.hc, k=3, db=kcs20, groups=1:2, colour.selected=TRUE)