This function returns a list (containing the order of nodes) travelled using a depth first search starting from the given node.
distal_to(
x,
node.idx = NULL,
node.pointno = NULL,
root.idx = NULL,
root.pointno = NULL
)Arguments
- x
A neuron
- node.idx, node.pointno
The id(s) of node(s) from which distal points will be selected.
node.idxdefines the integer index (counting from 1) into the neuron's point array whereasnode.pointnomatches the PointNo column which will be the CATMAID id for a node.- root.idx, root.pointno
The root node of the neuron for the purpose of selection. You will rarely need to change this from the default value. See
nodeargument for the difference betweenroot.idxandroot.pointnoforms.
Value
Integer 1-based indices into the point array of points that are distal to the specified node(s) when traversing the neuron from the root to that node. Will be a vector if only one node is specified, otherwise a list is returned
See also
Examples
# \donttest{
## Use EM finished DL1 projection neuron
## subset to part of neuron distal to a tag "SCHLEGEL_LH"
# nb distal_to can accept either the PointNo vertex id or raw index as a
# pivot point
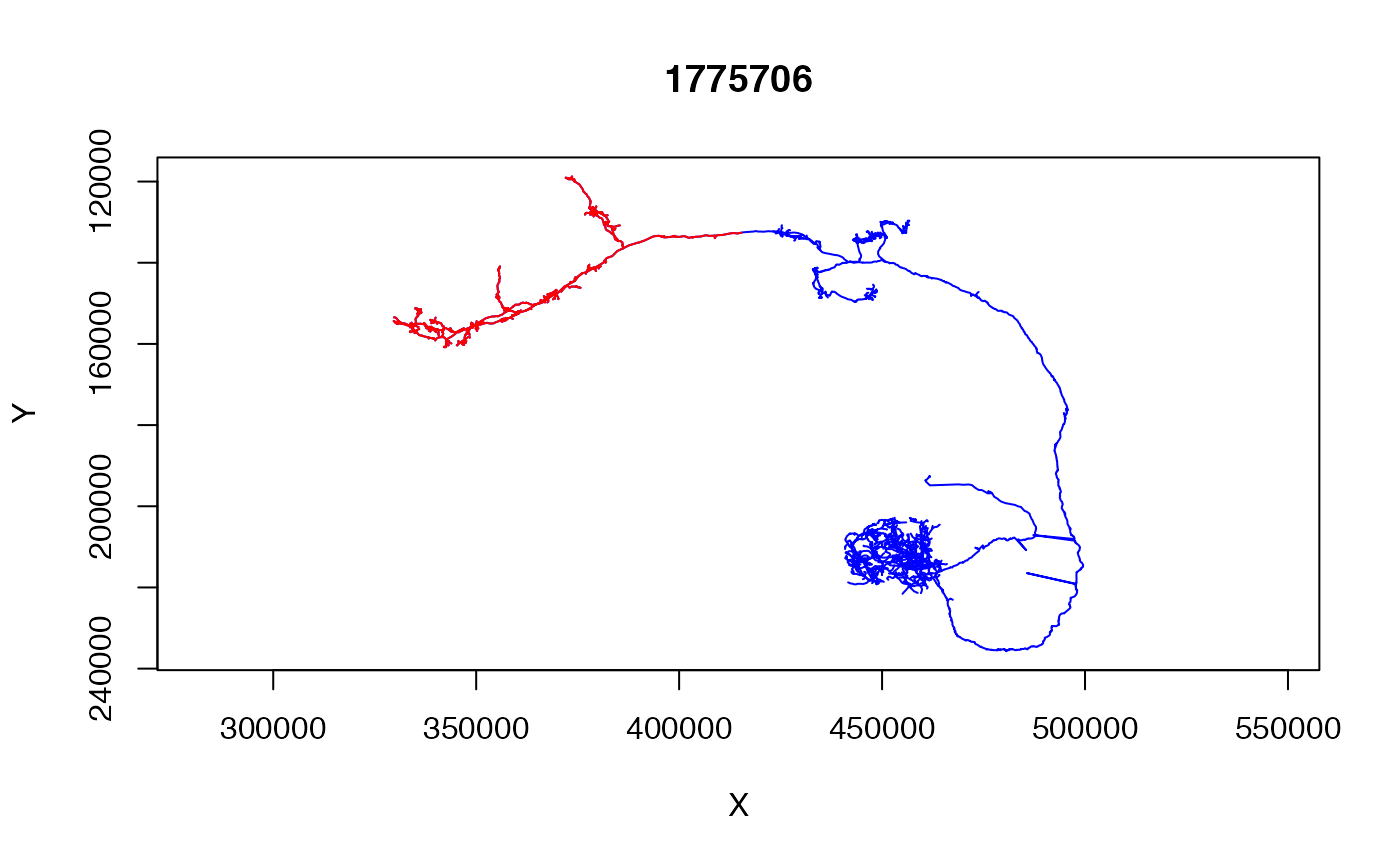
dl1.lh=subset(dl1neuron, distal_to(dl1neuron,
node.pointno = dl1neuron$tags$SCHLEGEL_LH))
plot(dl1neuron,col='blue', WithNodes = FALSE)
plot(dl1.lh, col='red', WithNodes = FALSE, add=TRUE)
 # }
# }