Meta data for hemibrain and flywire neurons
The package hemibrainr allows you to retrieve pre-computed/annotated meta data for thousands of neurons.
This includes neurons from both the hemibrain and
FlyWire ( FAFB ) connectomics data sets.
Unlike with more manual reconstruction environments like CATMAID, users concatenate volumetric segmentations to build neurons as 3D meshes. The segmentation was produced by the Seung group. With the package hemibrainr we can read flywire neurons as both meshes and skeletons. Meshes can be read directly from flywire, but skeletons need to be built from these meshes. Our package fafbseg can enable users to skeletonise neurons in R (e.g. fafbseg::skeletor), whereas hemibrainr exists to help users get hold of neuron data and match flywire neurons to hemibrain neurons.
Which is all useful stuff. To access some of this data, you will need access to the hemibrainr Google team drive. In order to connect R to this Google drive, you have a few options. Please see this article. The pipeline that produced this data can be found here. It is run nightly on a machine at the MRC LMB.
# Load package
remotes::install_github("natverse/hemibrainr")
library(hemibrainr)Hemibrain meta data
Hemibrain meshes
A mesh for the whole hemibrain volume is not available from neuPrint. We have made one. It is a 3D triangle mesh for the hemibrain surface as a .obj file. This mesh was generated by first merging individual ROI meshes from neuPrint and then filling the gaps in between in a semi-manual process.
nat::nopen3d() # holds right click to pan
plot3d(hemibrain.surf, col = "grey", alpha = 0.1) # plot the brain in raw voxel space
## plot3d(hemibrain_microns.surf, col = "pink", alpha = 0.1) # You can also plot the brain scaled to microns
hemibrain_view() # orients the brain to face forward. Note that hemibrain coordinate system has the anterior-posterior axis aligned with the Y axis (rather than the Z axis, which is more commonly observed)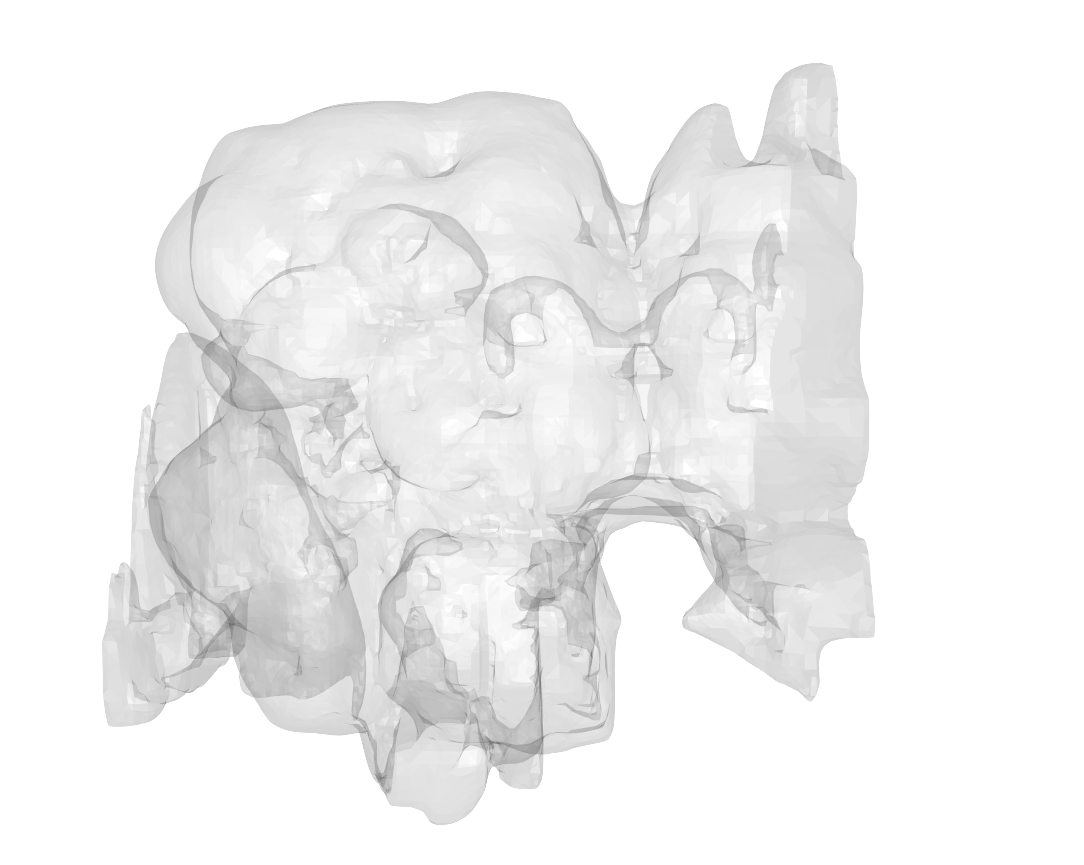
We have also generated 3D meshes of 51 olfactory + 7 thermo/hygrosensory antennal lobe glomeruli for the hemibrain volume. They have been made using antennal lobe projection neuron presynapses.
gloms = materials(hemibrain_al.surf)$name
for(glom in gloms){
plot3d(subset(hemibrain_al.surf,glom), add = TRUE, alpha = 0.3)
}
# Also exists, scaled to microns: hemibrain_al_microns.surf
# We have also stored meta-information rel;ated to these glomerular meshes
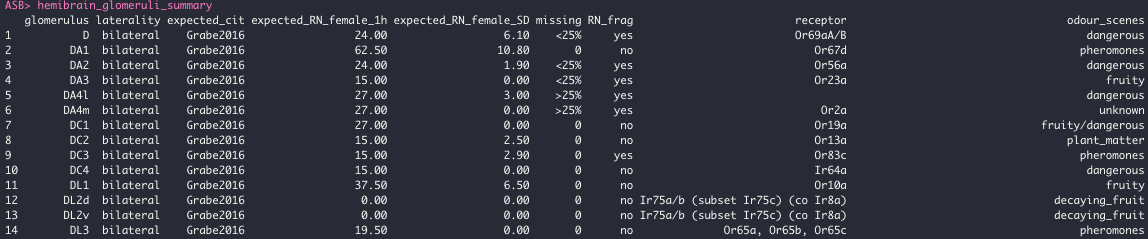
hemibrain_glomeruli_summary
## including whether or not they are cut-off in the hemibrain volumes
### And what sorts of odours seem to excite them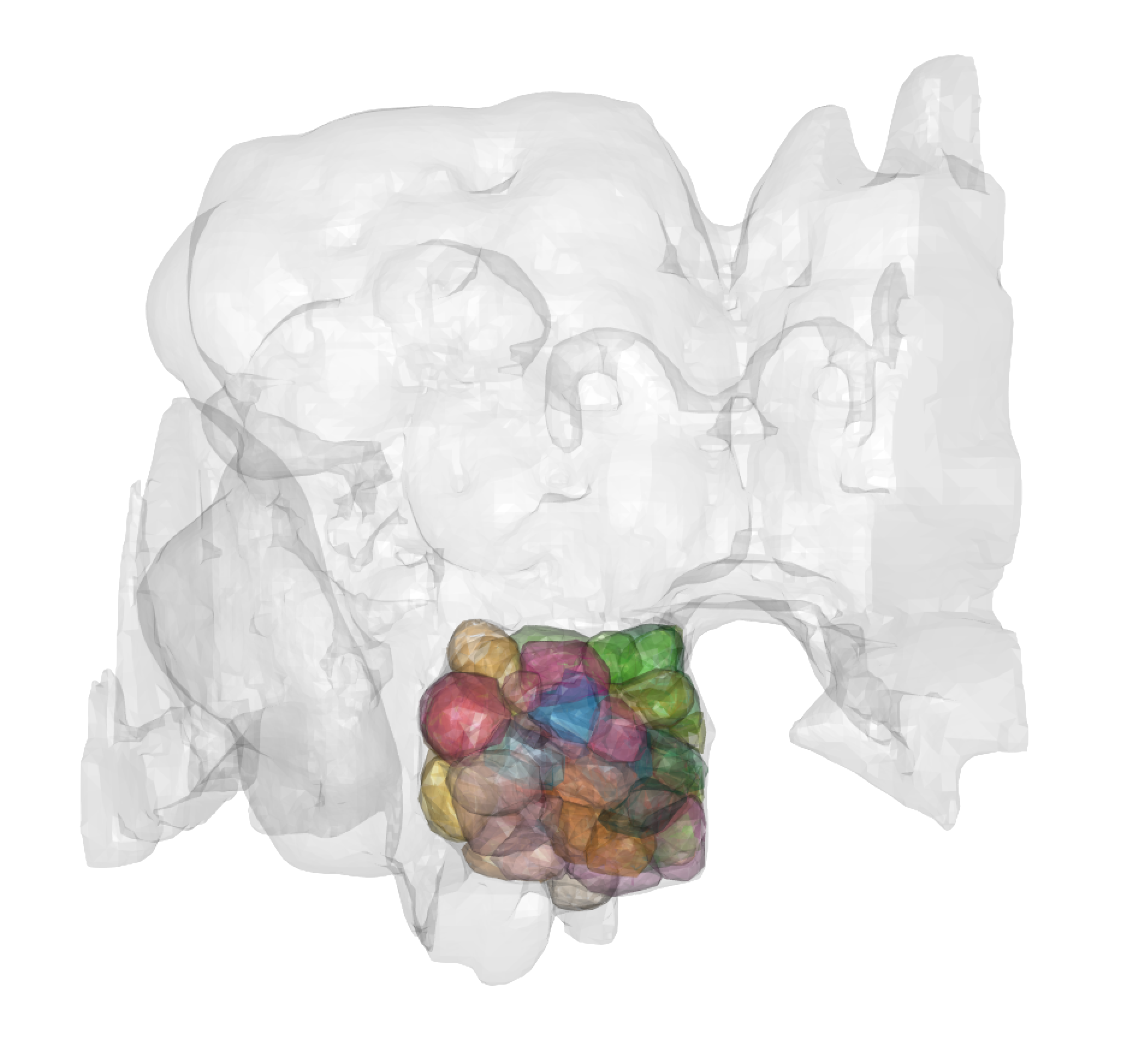
Hemibrain linage information
We have tried to map reported cell body fibres designations (Scheffer et al. 2020) and match them to hemilineage designations from the literature. Hemilineages are bundles of neurons that have developed together, and which likely express similar genes including those related to neurotransmitter expression (Lacin et al. 2019; Eckstein et al. 2020).
head(hemibrain_hemilineages)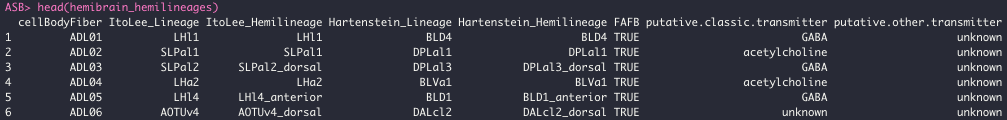
This mapping may be in precise, and we are working on creating a direct neuron-hemilineage mapping.
Hemibrain annotations for olfactory neurons
Many olfactory neurons have been annotated into their classes, cell types and olfactory layers in recent work Schlegel & Bates et al., 2021. This information is available in our packages as:
# These are all data frames available through the hemibrainr package:
head(rn.info) # meta data related to antennal lobe projection neurons
head(alln.info) # meta data related to antennal lobe projection neurons
head(pn.info) # meta data related to antennal lobe projection neurons
head(ton.info) # meta data related to antennal lobe projection neurons' targets
head(dn.info) # meta data related to antennal lobe projection neurons
?pn.info # get help information, including meaning of columns
# Includes things like cell types, connectivity types, lineage
# information, which olfactory layer a neuron belongs to
# (Schlegel and Bates 2021), putative transmitter, etc.
head(hemibrain_olfactory_layers) # layer information for every neuron in the hemibrain
# Dataset, as described in Schlegel and Bates et al. 2021.
We also provides groups of IDs for various neuron classes:
rn.ids # antennal lobe receptor neuron IDs
orn.ids # olfactory antennal lobe receptor neuron IDs
hrn.ids # thermo/hygrosensory antennal lobe receptor neuron IDs
pn.ids # antennal lobe projection neuron IDs
upn.ids # uniglomerular antennal lobe projection neuron IDs
mpn.ids # multiglomerular antennal lobe projection neuron IDs
vppn.ids # thermo/hygrosensory antennal lobe projection neuron IDs
dan.ids # mushroom body dopaminergic neuron neuron IDs
mbon.ids # mushroom body output neuron IDs
alln.ids # antennal lobe local neuron IDs
ton.ids # third-order olfactory neuron IDs
lhn.ids # lateral horn neurons IDs
dn.ids # decending neuron IDs (descend to ventral nervous system)
kc.ids # Kenyon cell IDs
apl.ids # APL IDs
cent.ids # LH centrifugal neuron IDs
lc.ids # lobular column visual projection neurons IDs