Here, our aim is to get the DM6 uniglomerular projection neuron, and obtain all of its synapses within the DM6 glomerulus. We then want to find all the partner neurons that connect with it, both up- and downstream, within that glomerulus.
# Load the libraries we need
library(hemibrainr)
library(fafbseg)
library(elmr)
library(ComplexHeatmap)
library(reshape2)
choose_segmentation('flywire')
# We need to use some python modules to skeletonise meshes
## Install them with this R code
# fafbseg::simple_python("full")
# We can query the virtual fly brain public CATMAID in order to obtain
# A CATMAID reconstruction of the DM6 uPN, and so find its flywire equivalent.
adult.conn = catmaid_login(server="https://catmaid-fafb.virtualflybrain.org/")
dm6.upn.ids.all = fafb14_to_flywire_ids("name:Uniglomerular mALT DM6 adPN")
### Includes both left and right uPNs
# You can now look at all fo the IDs in flywire, and connect
# Them up if needed. Some will be wrong. Look at the hits column, probably
# Best to just look through things with high hits.
# This now gives you one flywire ID per neuron
dm6.upn.ids.biggest = fafb14_to_flywire_ids("name:Uniglomerular mALT DM6 adPN",
only.biggest = TRUE)
dm6.upn.ids = dm6.upn.ids.biggest$root_id
# Get meshes
dm6.upn.meshes = fafbseg::read_cloudvolume_meshes(dm6.upn.ids)
# Look good?
nopen3d()
plot3d(dm6.upn.meshes[1], alpha = 0.5, col = "red")
plot3d(FAFB14, col = "grey", alpha = 0.1)
# Let us get the skeletons
dm6.upn.skels = fafbseg::skeletor(dm6.upn.ids, method = 'wavefront')
### If this crashes, follow these instructions instead: http://natverse.org/fafbseg/articles/articles/installing-cloudvolume-meshparty.html
### Or cheat and just get the skeletons from the drive: dm6.upn.skels = subset(flywire_neurons(), root_id %in% dm6.upn.ids)
# Look good?
nopen3d()
plot3d(dm6.upn.skels, lwd = 3)
plot3d(FAFB14, col = "grey", alpha = 0.1)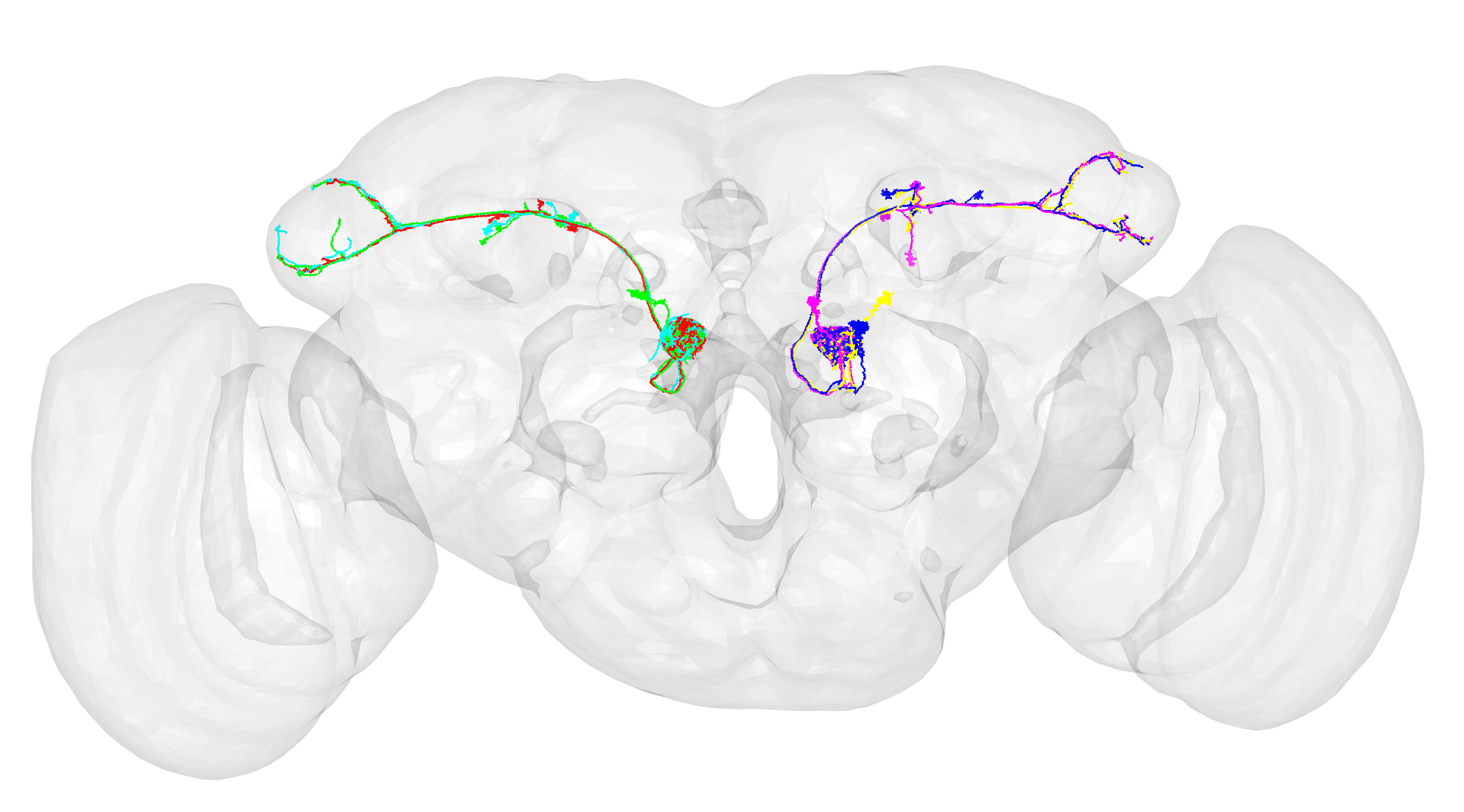
# Now let us attach the synapses with prediction
dm6.upn.syn = flywire_neurons_add_synapses(dm6.upn.skels,
cleft.threshold = 50,
transmitters = FALSE,
Verbose = TRUE,
OmitFailures = TRUE,
.parallel = FALSE)
# Now split into axon and dendrite
dm6.upn.flow = hemibrainr::flow_centrality(dm6.upn.syn, mode = "centrifugal", polypre = TRUE, split = "synapses", .parallel = FALSE, OmitFailures = TRUE)
# Plot together
nopen3d()
plot3d(FAFB14, col = "grey", alpha = 0.1)
plot3d_split(dm6.upn.flow)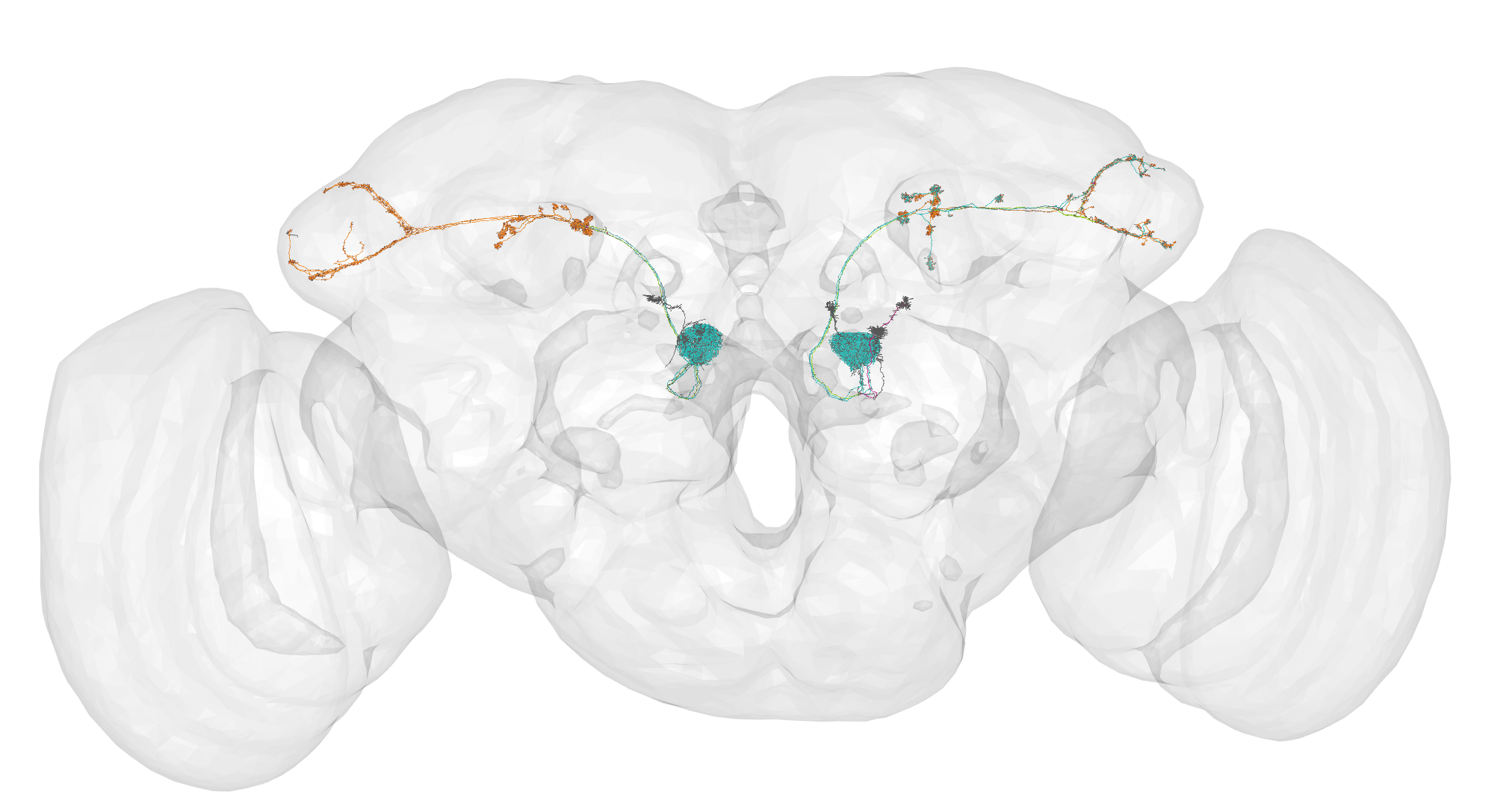
# Get synapse data
dm6.upn.synapses = hemibrain_extract_synapses(dm6.upn.flow, .parallel = FALSE, OmitFailures = FALSE)
# Obtain partner neurons, but only at the dendrite
dm6.upn.synapses.dend = subset(dm6.upn.synapses, Label == "dendrite")
dm6.upn.synapses.dend = subset(dm6.upn.synapses.dend, y > 175000)
# See the DM6 glomerulus
dm6 = subset(flywire_al.surf, 'DM6')
nopen3d()
plot3d(FAFB14, alpha = 0.1)
plot3d(dm6, col = hemibrain_bright_colors["cerise"], alpha = 0.3)
points3d(nat::xyzmatrix(dm6.upn.synapses.dend), col = hemibrain_bright_colors["marine"])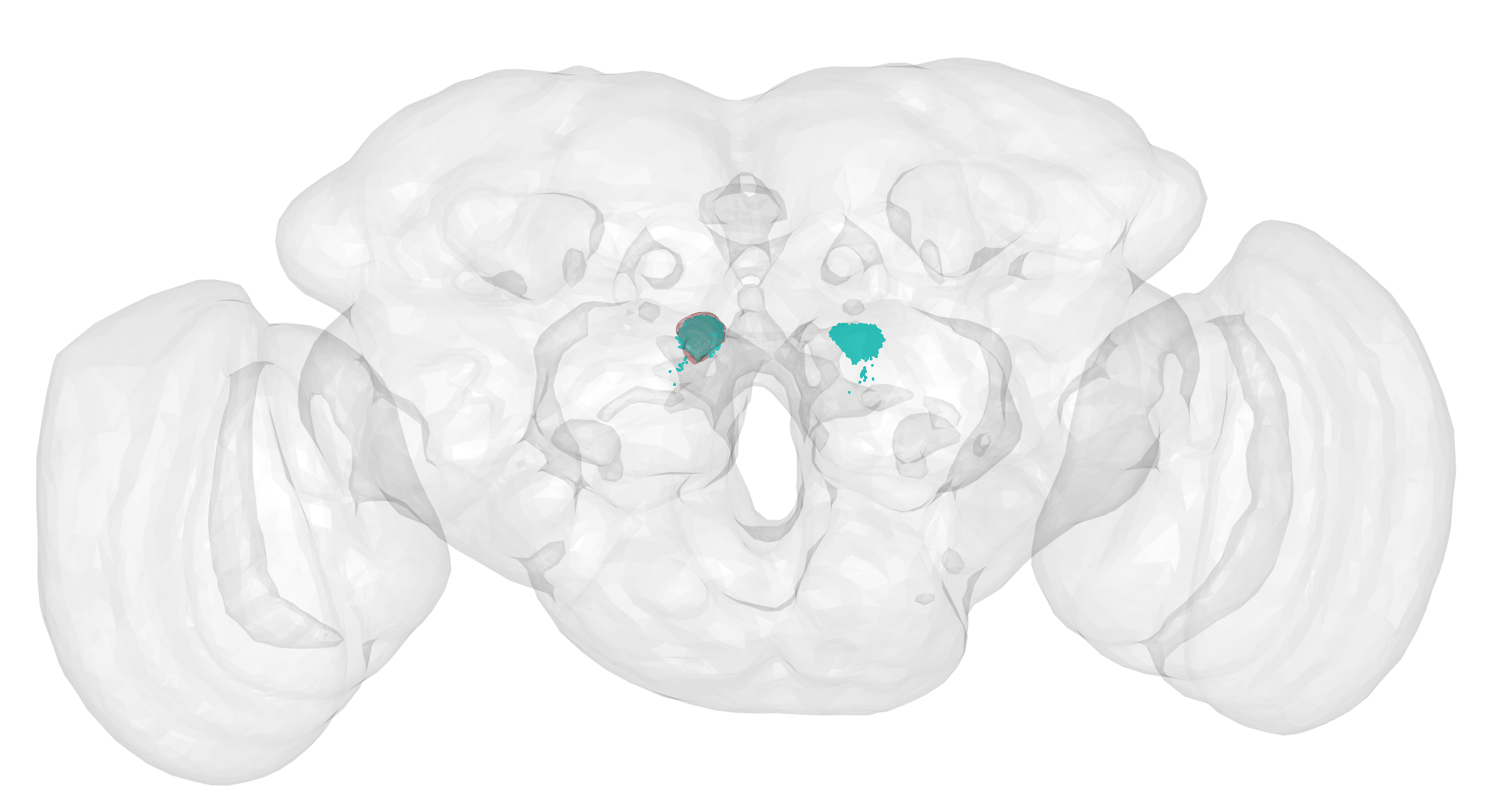
# Get connections in an edgelist
dm6.upn.conns = hemibrain_extract_connections(dm6.upn.synapses.dend)
dm6.upn.conns.strong = subset(dm6.upn.conns, count >= 10)
dm6.upn.partner.ids = unique(as.character(dm6.upn.conns.strong$partner))
dm6.upn.partner.skels = fafbseg::skeletor(dm6.upn.partner.ids, method = 'wavefront')
## These IDs constitute the neurons you should examine and 'clean up' in flywire
# Treat GABA/Glut synapses as inhibitory (if you have the predicted transmitters)
dm6.upn.conns.strong$count[dm6.upn.conns.strong$top_nt%in%c("gaba","glutamate")] = -1*dm6.upn.conns.strong$count[dm6.upn.conns.strong$top_nt%in%c("gaba","glutamate")]
# Plot!
nopen3d()
plot3d(FAFB14, alpha = 0.1)
plot3d(dm6.upn.partner.skels, soma = 5000)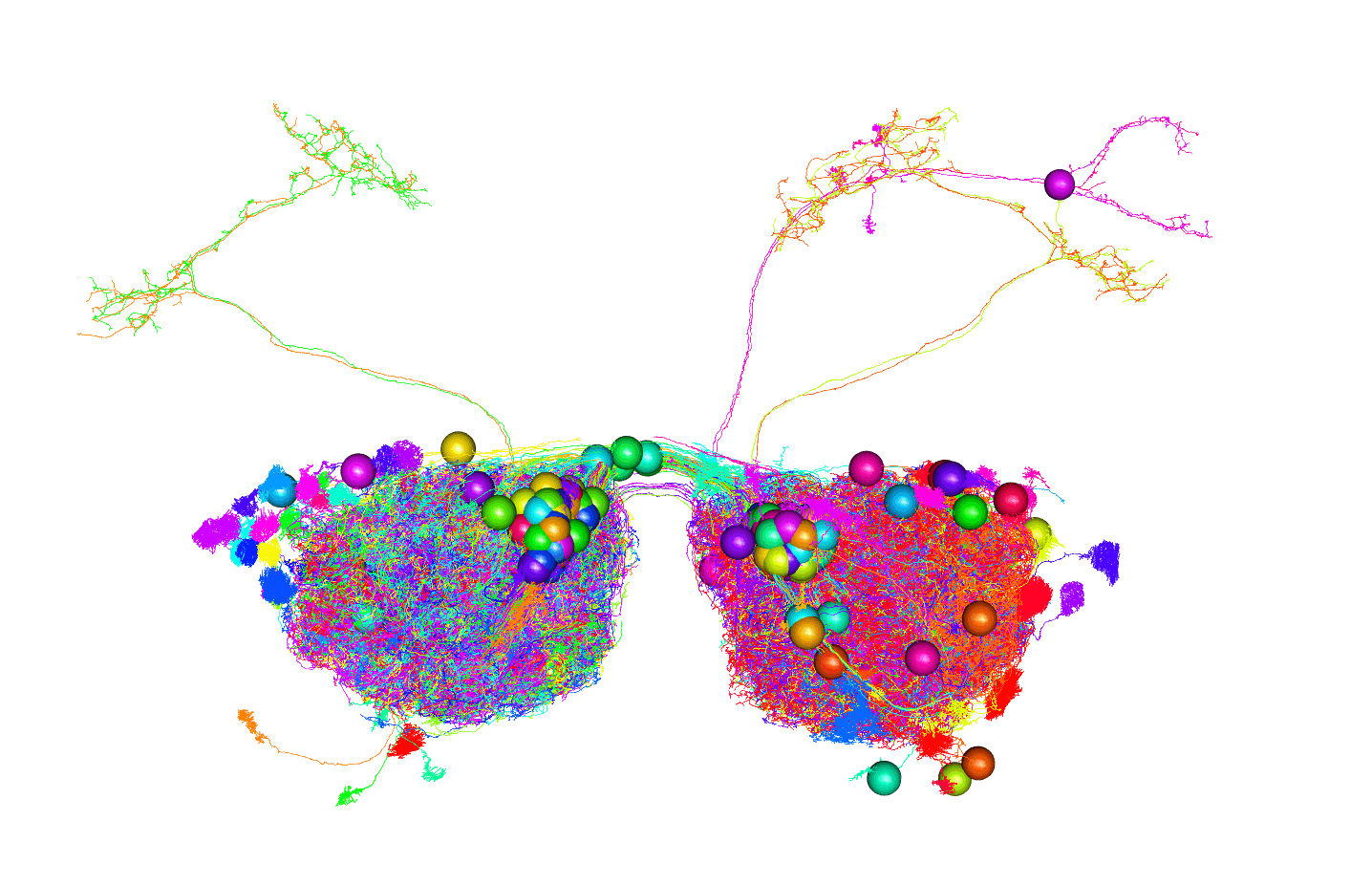
# Get up- and downstream connectivity matrices
dm6.conn.mat.downstream = acast(data = subset(dm6.upn.conns.strong, prepost == 1),
formula = root_id ~ partner,
fun.aggregate = sum,
value.var = "count",
fill = 0)
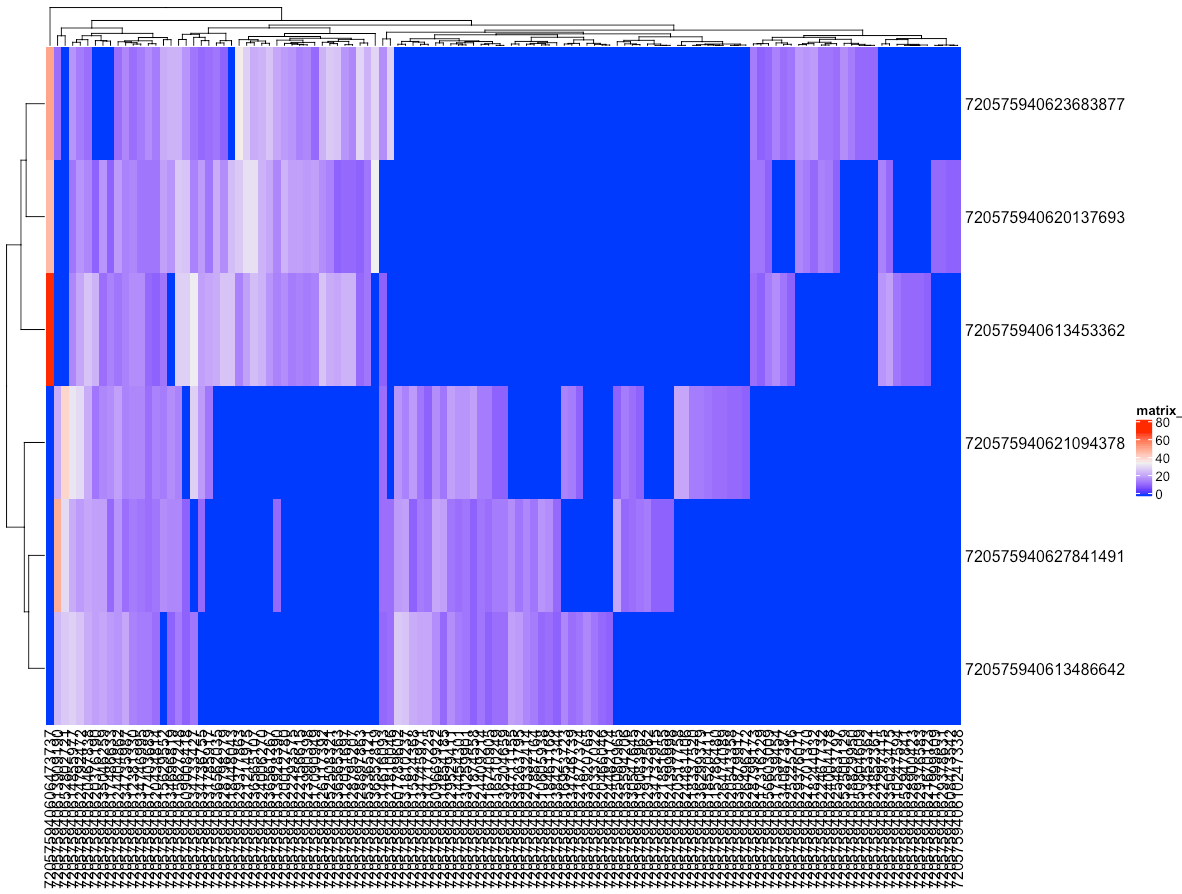
ComplexHeatmap::Heatmap(dm6.conn.mat.downstream)
dm6.conn.mat.upstream = acast(data = subset(dm6.upn.conns.strong, prepost == 0),
formula = root_id ~ partner,
fun.aggregate = sum,
value.var = "count",
fill = 0)
ComplexHeatmap::Heatmap(dm6.conn.mat.upstream)