“This history of civilization proves beyond doubt just how sterile the repeated attempts of metaphysics to guess at nature’ s laws have been. Instead, there is every reason to believe that when the human intellect ignores reality and concentrates within, it can no longer explain the simplest inner workings of life’ s machinery or of the world around us.” (Santiago Ramón y Cajal
Axons, dendrite and other compartments
The philosophy of hemibrainr is to try and treat axons and dendrites separately, where we can. Generally, the division between axon and dendrite is ignored when people use graph theoretic approach on connectome data, or otherwise build their models. However, dendrite and axons may function differently - we just don’t yet know in insects, how differently.
Other compartments include the ‘primary neurite’ or ‘cell body fibre’ (purple, below) (Ito et al., 2014), the non-synaptic cable that attaches the neuron’s cell body, its ‘soma’ to the rest of the neuron. There is also a ‘primary dendrite’, or ‘linker’ (green, below), which is non-synaptic cable that connects a neuron’s dendrite and axon, through which an action potential presumably travels.
Splitting a neuron
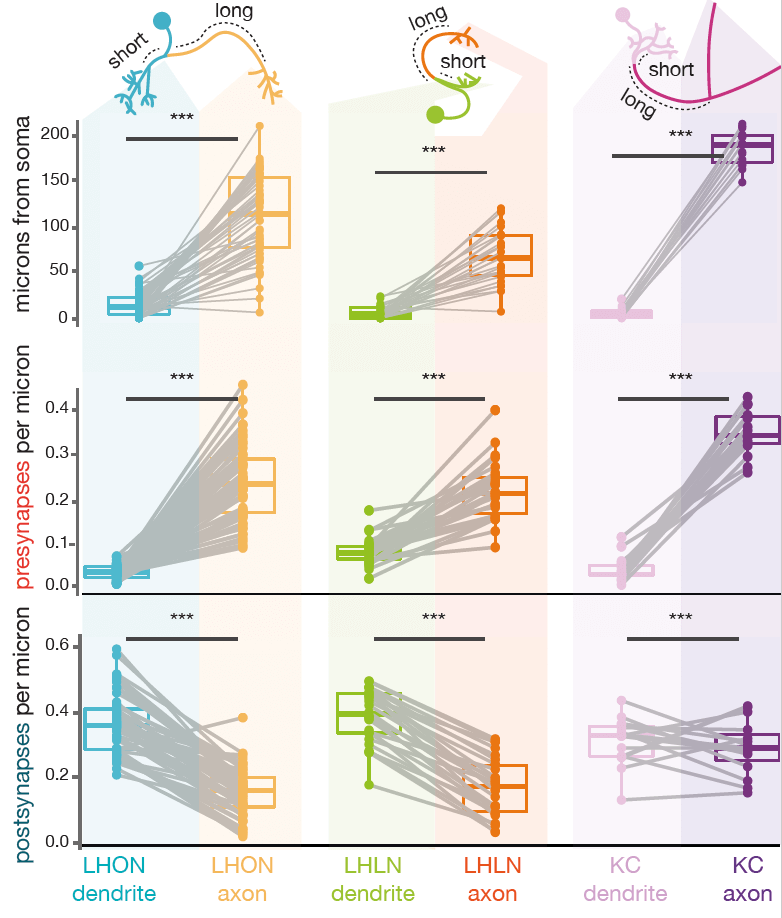
Neurons can be split into a principle input zone, a dendrite, and a principle output zone, an axon. Dendrites tend to be more spindly and axons bulkier, with boutons packed with presynapses, ready to release neurotransmitter.
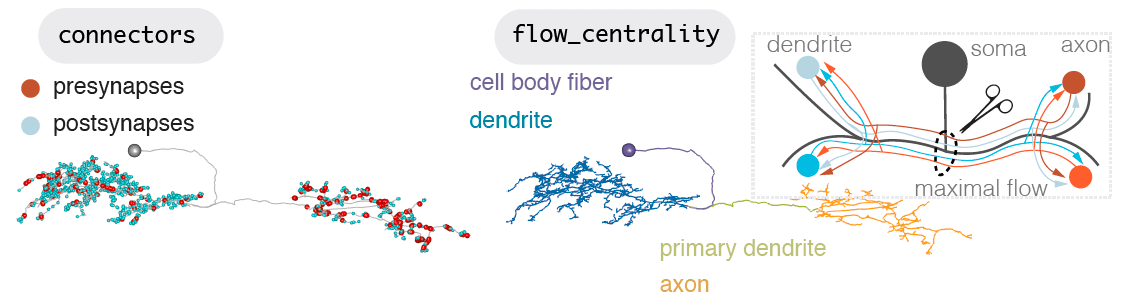
But axons and dendrites are not a totally clear division for most neurons. A neuron may have more than one of each. It may also be more ‘amacrine-like’ and lack this division, though this is less common. Dendrites can have outputs, and axons can have inputs. Axons may connect with axons, even dendrites may input and axon.
However, all fly neurons examined can be split into an axon-like and a dendrite-like compartment (Schneider-Mizell et al., 2016)). The density of outputs in the axon is higher than in the dendrite. The density of inputs to the dendrite, are higher than in the axon (Bates & Schlegel et al., 2020)). Interestingly, the ‘start’ of a dendrite is almost always closer to the neuron’s cell body than the start of its axon. See our recent data (Bates & Schlegel et al., 2020)) for three broad classes of olfactory neuron:
The biophysical impact of axo-axonic connections is unclear – they can have non-intuitive effects depending on the timing of action potentials in connected partners (Burrows and Laurent, 1993; Burrows and Matheson, 1994; Cuntz et al., 2007; Haag and Borst, 2004).
Splitting a neuron by flow centrality
We can split neurons algorithmically by calculating a neuron’s ‘flow centrality’ (Schneider-Mizell et al., 2016)).
From (Schneider-Mizell et al., 2016)): > “We use flow centrality for four purposes. First, to split an arbor into axon and dendrite at the maximum centrifugal SFC, which is a preliminary step for computing the segregation index, for expressing all kinds of connectivity edges (e.g. axo-axonic, dendro-dendritic) in the wiring diagram, or for rendering the arbor in 3d with differently colored regions. Second, to quantitatively estimate the cable distance between the axon terminals and dendritic arbor by measuring the amount of cable with the maximum centrifugal SFC value. Third, to measure the cable_length of the main dendritic shafts using centripetal SFC, which applies only to insect neurons with at least one output syn- apse in their dendritic arbor. And fourth, to weigh the color of each skeleton node in a 3d view, providing a characteristic signature of the arbor that enables subjective evaluation of its identity.”
Explore their methods for more information.
Tutorial
First, let us get and load the packages we will need:
# install
if (!require("remotes")) install.packages("remotes")
remotes::install_github("natverse/hemibrainr")
# use
library(hemibrainr)Split neurons from the hemibrain
Let us examine some third-order olfactory neurons (TONs), which we have stored in hemibrainr as hemibrainr::ton.ids.
# Let's examine a random set of tons
ids = sample(hemibrainr::ton.ids,10)
# Read neurons from neuprint
neurons = neuprintr::neuprint_read_neurons(ids)
# Now make sure the neurons have a soma marked
## Some hemibrain neurons do not, as the soma was chopped off
neurons.checked = hemibrain_skeleton_check(neurons, meshes = hemibrain.surf)
# Split neuron
## These are the recommended parameters for hemibrain neurons
neurons.flow = flow_centrality(neurons.checked,
polypre = TRUE,
mode = "centrifugal",
split = "synapses")
## What has this returned?
## It is still a neuronlist object, but now more infromation has been added
## To each neuron in the neuron list. If neuron = neurons.flow[[1]], then
## the neuron has flow centrality information added to neuron$d and a segregation index score.
## The neuron$d$Label now gives the compartment,
## where axon is Label = 2, dendrite Label = 3, primary dendrite Label = 9
## and primary neurite Label = 7. Soma is Label = 1.
## (note, some outputs from various function give names rather than numbers)
## We can see this, as so:
neuron = neurons.flow[[1]]
head(neuron$d)
### standard_compartments is a helper function to change numbers to words
### for the different compartment types.However, to make things easier, we can do this all in one step:
# Get the split neurons
neurons.flow = hemibrain_read_neurons(ids, remote = FALSE)
## This function reads neurons using neuptinr
## If remote == TRUE, then neurons read from linked google drive. See relevant article for more information.Get already split neurons
Splitting neurons can take a little time. We can also load thousands of pre-split neurons from the hemibrainr Google team drive. In order to connect R to this Google drive, you have a few options. Please see this article. Once you have access and have mounted the drive, you should be able to do:
# All split hemibrain neurons
db = hemibrain_neurons()
length(db)
## This is a nat::neuronlistfh object.
### This means that a data frame for the neurons and information
### specifying where to find each neuron's data is read into R -
### but not the whole, huge nat::neuronlist object. This saves on memory
### for your R session. When an operation that requires the actual
### neuron data is performed, neurons are read into R.
# Or just load the neurons you want
neurons.flow = hemibrain_read_neurons(ids, remote = TRUE)Hemibrain select cable
If we want, we can select just the cable and synapses that constitute one of the neuron’s compartments.
For example:
# Extract individual bits of cable
axons = axonic_cable(neurons.flow)
dendrites = dendritic_cable(neurons.flow)
pnts = primary_neurite(neurons.flow)
pds = primary_dendrite_cable(neurons.flow)
tracts = tract_cable(neurons.flow)
# Plot
nat::nopen3d()
hemibrain_view()
plot3d(axons, col = "orange")
plot3d(dendrites, col = "cyan")
plot3d(pnts, col = "purple", soma = 500)
plot3d(pds, col = "green")
plot3d(tracts, col = "black", lwd = 5)
# We can see, for example, just axon output synapses:
axon.synapses = hemibrain_extract_synapses(axons, prepost = "PRE")
head(axon.synapses)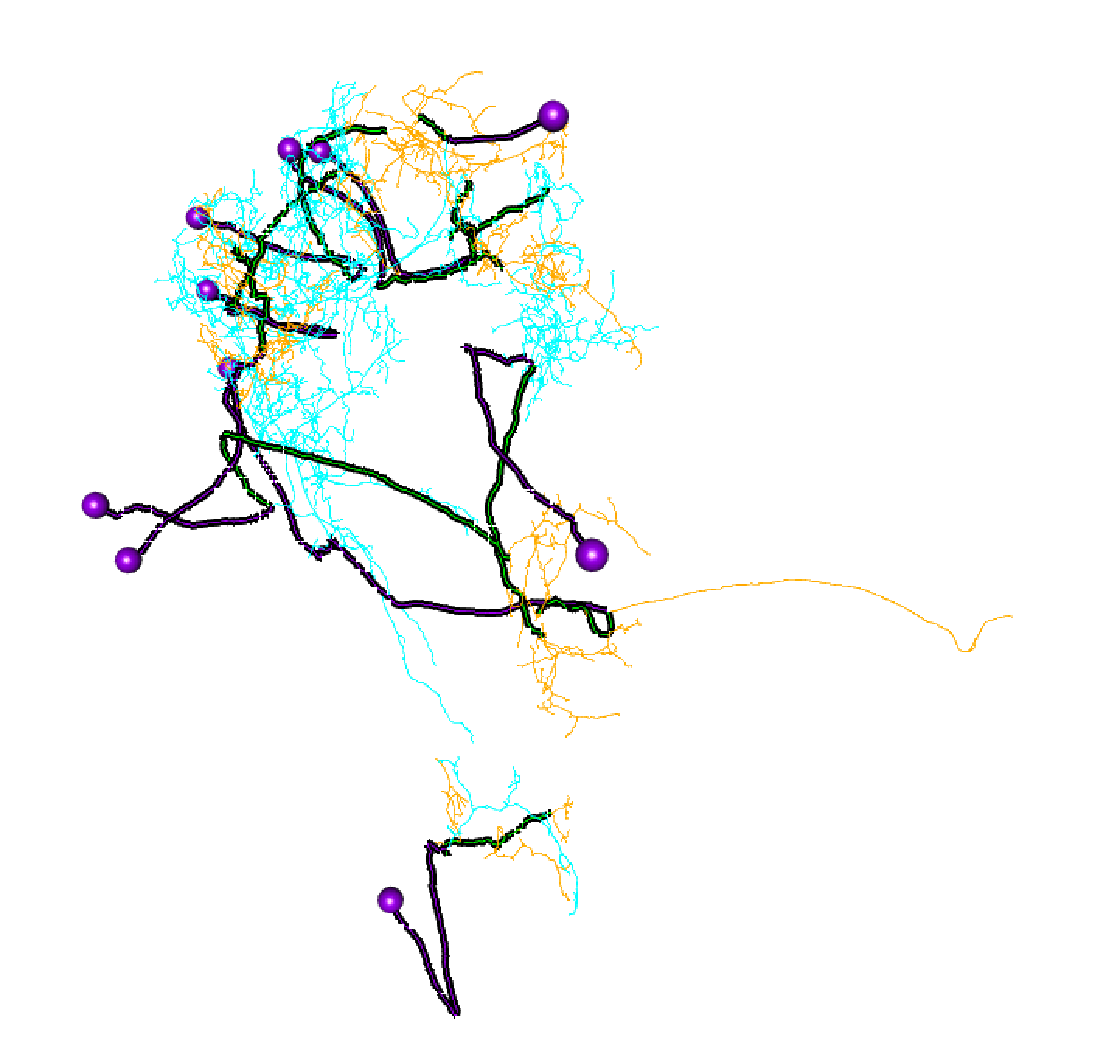
Visualise the split
We can see the neruon splits, using hemibrainr::plot3d_split:
# Plot 3D
nat::nopen3d()
hemibrain_view()
plot3d_split(neurons.flow)
plot3d(hemibrain.surf, col = "lightgrey", alpha = 0.1, add = TRUE)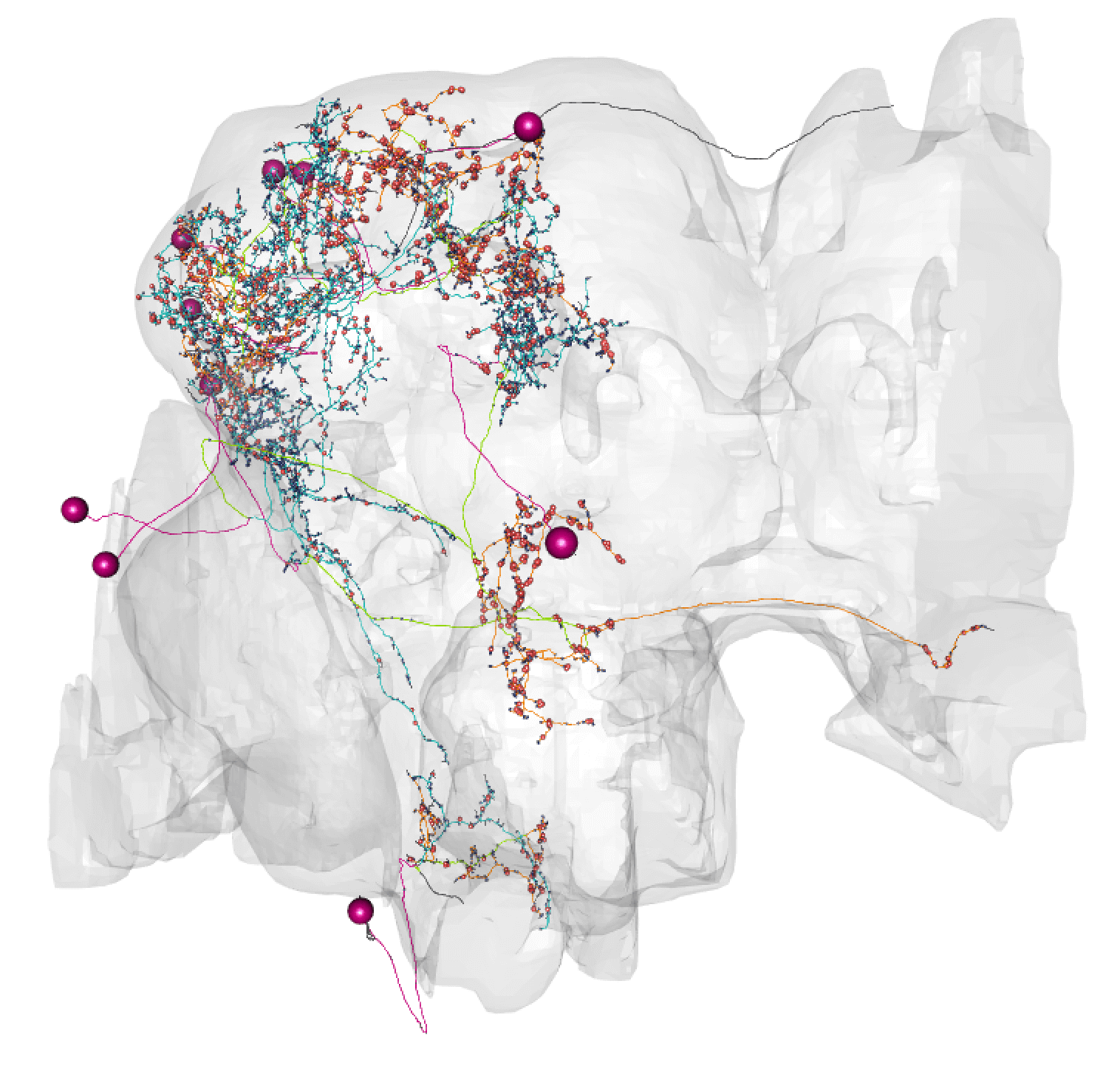
Here, lines: blue = dendrite, orange = axon, green = linker, purple = cell body fibre.
Balls: red = presynapse (output), navy blue = postsynapse (input), purple = soma.
We can also scan through split to see them individually, using hemibrainr::nlscan_split:
# Plot 3D
clear3d()
nlscan_split(neurons.flow)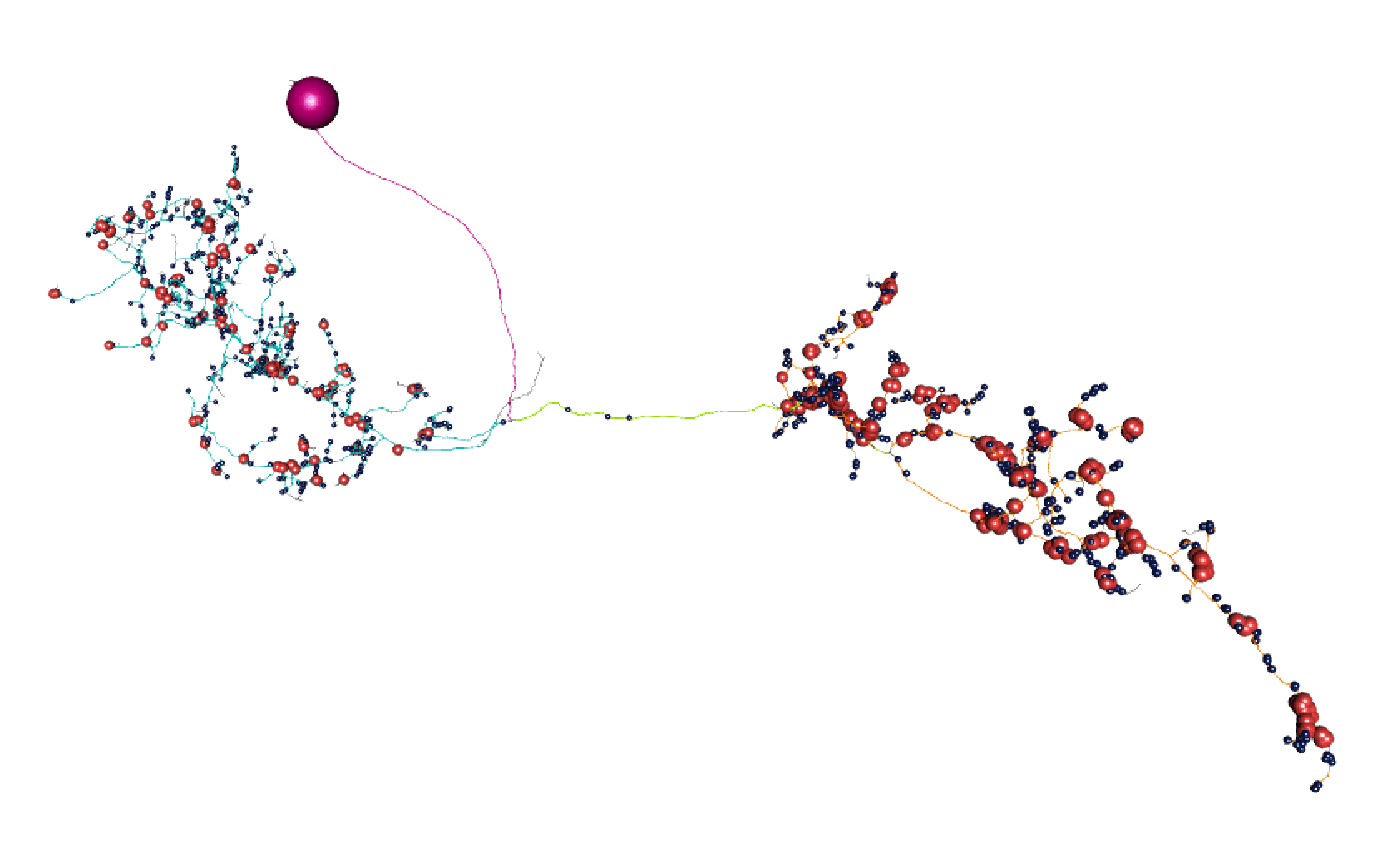
So I think most of those splits are pretty convincing.
Split connectivity
It will likely be important, when looking at neuron connectivity, to know whether we are examining a axo-dendritic, axo-axonic, dendro-dendritic, etc., connection.
We can see where synapses on these neurons lie, using:
neuron.synapses = hemibrain_extract_synapses(neurons.flow, prepost = "BOTH")
table(neuron.synapses$prepost, neuron.synapses$Label)Here, there is one entry per synapse. The prepost column tells us whether the synapse is presynaptic (0, an output synapse) or postsynaptic (1, an input synapse).
We can also easily get one entry per target-source connection, by:
# Extract connections
neuron.connections = hemibrain_extract_connections(neurons.flow, prepost = "BOTH")
neuron.connections.strong = subset(neuron.connections, count >= 5)
# Connections from neurons' dendrites
hist(subset(neuron.connections.strong, Label == "dendrite" & prepost == 1)$count,
main="Connections from neurons' dendrites",
xlab="Strength of neuron->partner synaptic connection",
col="cyan",
freq=TRUE,
breaks = max(subset(neuron.connections.strong, Label == "dendrite")$count)
)
# Connections from neurons' axons
hist(subset(neuron.connections.strong, Label == "axon" & prepost == 1)$count,
main="Maximum daily temperature at La Guardia Airport",
xlab="Strength of neuron->partner synaptic connection",
col="orange",
freq=TRUE,
breaks = max(subset(neuron.connections.strong, Label == "axon")$count)
)Here, the prepost column tells us whether the partner is presynaptic (0, upstream source) or postsynaptic (1, downstream target).
We might also want to know the ‘Label’ of the partner cell. We can get an edgelist with this information:
## The function hemibrain_extract_compartment_edgelist will give an edgelist
## for connections between the neruons you provide to it.
## To make sure some of our randomly selected neurons connect
## Let's also grab some PNs
pns.flow = hemibrain_read_neurons(hemibrainr::upn.ids, remote= TRUE)
### Set remote to FALSE, if you do not have the google drive mounted.
# Extract connections
pns.elist = hemibrain_extract_compartment_edgelist(pns.flow, .progress = 'text')
pns.elist.strong = subset(pns.elist, count >= 10 | norm > 0.01)
sort(table(pns.elist.strong$connection))
## A lot of axo-axonic and dendro-dendritic action amongst the PNsHere, pre is the bodyid of the presynaptic (upstream source) neuron and post is the bodyid of the postsynaptic (downstream target) neuron. The norm column gives a synaptic weight normalised by the total_inputs of the downstream neuron (post), i.e. count/total postsynapses on ‘post’.
Please see this article to see a more in-depth example of how to efficiently work neuron connectivity data from hemibrainr.