Get connectivity between ROIs (summary or data frame of connecting neurons)
Source:R/roi.R
neuprint_ROI_connectivity.RdGet connectivity between ROIs (summary or data frame of connecting neurons)
neuprint_ROI_connectivity(
rois,
full = TRUE,
statistic = c("weight", "count"),
cached = !full,
dataset = NULL,
conn = NULL,
...
)Arguments
- rois
regions of interest for a dataset
- full
return all neurons involved (TRUE, the default) or give a numeric ROI summary (FALSE)
- statistic
either "weight" or count" (default "weight"). Which number to return (see neuprint explorer for details) for summary results (either (when
full=FALSE)- cached
pull precomputed results (TRUE) or ask server to recalculate the connectivity (FALSE). Only applicable to summary results when
full=FALSE.- dataset
optional, a dataset you want to query. If
NULL, the default specified by your R environ file is used or, failing that the current connection, is used. Seeneuprint_loginfor details.- conn
optional, a neuprintr connection object, which also specifies the neuPrint server. If NULL, the defaults set in your
.Rprofileor.Renvironare used. Seeneuprint_loginfor details.- ...
methods passed to
neuprint_login
Details
When requesting summary connectivity data between ROIs, we recommend
setting cached=FALSE. We have noticed small differences in the
connections weights, but computation times can get very long for more than
a handful of ROIs.
Examples
# \donttest{
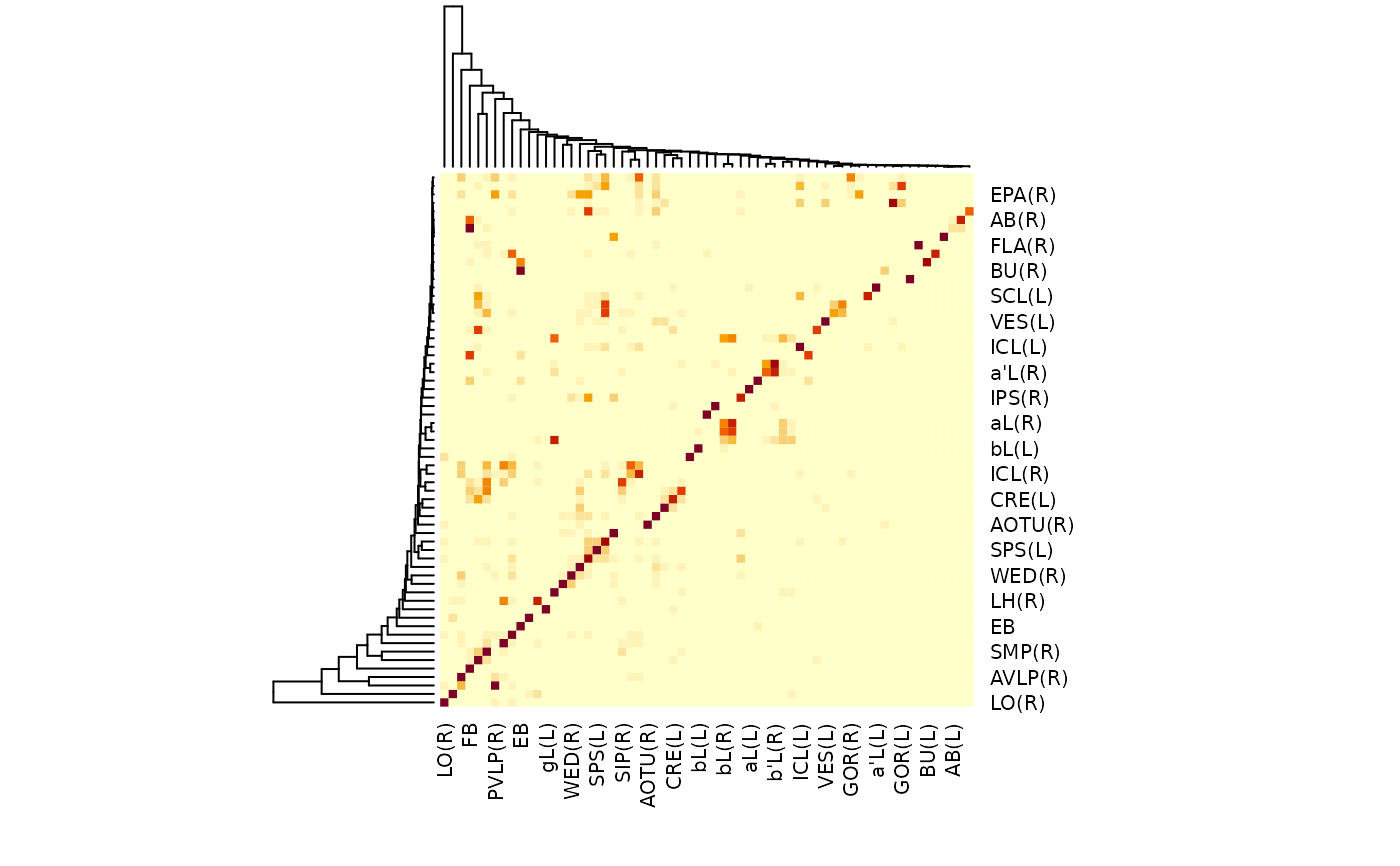
aba <- neuprint_ROI_connectivity(neuprint_ROIs(superLevel = TRUE),full=FALSE)
heatmap(aba)
 # }
# }