Finding neurons with multiple somata
Gregory Jefferis
2020-06-08
Source:vignettes/articles/multiple_somata.Rmd
multiple_somata.RmdIntro
Occasionally CATMAID users add the soma tag multiple times in error. We can find such neurons and generate CATMAID URLs to inspect them.
Finding neurons with multiple somata
First let’s fetch information about all the labels (aka tags) applied to nodes in the current project.
label_stats=catmaid_get_label_stats()
Now let’s restrict to cases where there are multiple soma tags per skeleton
# select soma labels soma_labels = label_stats %>% filter(labelName == 'soma') %>% group_by(skeletonID) # select skeleton ids for neurons with multiple cell bodies multiple_soma = soma_labels %>% count(skeletonID) %>% filter(n > 1) %>% arrange(desc(n)) multiple_soma_info = soma_labels %>% filter(skeletonID %in% multiple_soma$skeletonID) multiple_soma_info = soma_labels %>% filter(skeletonID%in% multiple_soma$skeletonID)
XYZ position of the nodes we picked
# wrapper for tree node details node_details <- function(tnid) { res=catmaid_get_treenodes_detail(tnid) ul=catmaid_get_user_list() res$login=ul$login[match(res$user_id, ul$id)] # we expect these to be capitalised elsewhere cn=colnames(res) cn[cn%in%c("x","y","z")]=toupper(cn[cn%in%c("x","y","z")]) colnames(res) <- cn res[c("X","Y","Z","radius","login")] } # note that we need to transpose the results of vnode_xyz to get X,Y,Z columns multiple_soma_info <- cbind(as.data.frame(multiple_soma_info), node_details(multiple_soma_info$treenodeID))
Now let’s calculate the distance from the neuropil surface
multiple_soma_info$d=pointsinside(xyzmatrix(multiple_soma_info), FAFB.surf, rval = 'distance') multiple_soma_info %>% arrange(skeletonID, d) %>% group_by(skeletonID) %>% mutate(rank=row_number()) -> multiple_soma_info kable(multiple_soma_info)
| labelID | labelName | skeletonID | treenodeID | X | Y | Z | radius | login | d | rank |
|---|---|---|---|---|---|---|---|---|---|---|
| 2773 | soma | 1298891 | 7525237 | 395632.7 | 95574.06 | 177520 | 2855 | batesa | -9679.5078 | 1 |
| 2773 | soma | 1298891 | 59494856 | 404913.9 | 96547.42 | 161000 | 51 | serratosal | 5889.5156 | 2 |
| 2773 | soma | 2015619 | 56565186 | 322684.0 | 154162.50 | 201600 | 1985 | serratosal | -13418.5234 | 1 |
| 2773 | soma | 2015619 | 18010667 | 371781.5 | 128202.72 | 176640 | -1 | batesa | 14364.6289 | 2 |
| 2773 | soma | 3658067 | 12345094 | 618823.6 | 218756.61 | 60680 | 2440 | alghailanis | 2672.3125 | 1 |
| 2773 | soma | 3658067 | 30290984 | 413013.0 | 240218.00 | 145680 | -1 | polskyj | 13872.9307 | 2 |
| 2773 | soma | 3721315 | 12460766 | 434532.3 | 254146.14 | 84880 | 3918 | calles | -1958.2344 | 1 |
| 2773 | soma | 3721315 | 12459557 | 433765.5 | 253732.36 | 84320 | -1 | dacksa | -1411.6875 | 2 |
| 2773 | soma | 3726283 | 35469519 | 322771.5 | 155343.50 | 181680 | 2093 | tenshawe | -4883.0469 | 1 |
| 2773 | soma | 3726283 | 35469478 | 323028.0 | 155599.00 | 181200 | -1 | tenshawe | -4637.3281 | 2 |
| 2773 | soma | 3788293 | 19104385 | 370375.0 | 152519.00 | 215520 | 1904 | batesa | -7053.4844 | 1 |
| 2773 | soma | 3788293 | 19104384 | 370363.0 | 152491.00 | 215400 | 1889 | batesa | -6953.6719 | 2 |
| 2773 | soma | 4705224 | 61576868 | 333411.7 | 155215.83 | 196000 | 1791 | serratosal | -2653.0000 | 1 |
| 2773 | soma | 4705224 | 21007576 | 381708.0 | 129635.00 | 171280 | -1 | robertsr | 17971.9297 | 2 |
| 2773 | soma | 6645033 | 6781342 | 617127.0 | 261972.00 | 67160 | 3076 | calles | 226.7500 | 1 |
| 2773 | soma | 6645033 | 41219199 | 495504.6 | 264349.25 | 62520 | -1 | coatesk | 11138.5625 | 2 |
| 2773 | soma | 6645033 | 43899462 | 484171.5 | 264798.03 | 77240 | -1 | dacksa | 19888.3066 | 3 |
| 2773 | soma | 7145440 | 61582172 | 334965.7 | 152653.12 | 194360 | 2081 | serratosal | 352.7344 | 1 |
| 2773 | soma | 7145440 | 23455383 | 358511.0 | 139952.00 | 190680 | -1 | batesa | 10689.1250 | 2 |
| 2773 | soma | 7937976 | 57573136 | 320074.8 | 162848.39 | 185360 | 1702 | serratosal | -8637.9688 | 1 |
| 2773 | soma | 7937976 | 13380406 | 377685.2 | 210574.42 | 176800 | -1 | batesa | 4119.2852 | 2 |
| 2773 | soma | 8758601 | 49116764 | 440101.5 | 203967.33 | 56360 | 2454 | helmickl | 988.2656 | 1 |
| 2773 | soma | 8758601 | 52299679 | 476734.8 | 214770.89 | 39800 | -1 | helmickl | 11180.4805 | 2 |
| 2773 | soma | 9623317 | 32110234 | 428951.1 | 298439.03 | 209400 | 1781 | eichlerk | -8578.3359 | 1 |
| 2773 | soma | 9623317 | 59058312 | 497413.8 | 339922.00 | 164720 | 42 | eichlerk | 19744.7188 | 2 |
| 2773 | soma | 10108062 | 33531079 | 419915.0 | 224525.00 | 51080 | -1 | coatesk | -3335.2869 | 1 |
| 2773 | soma | 10108062 | 40684585 | 513639.0 | 219860.00 | 46640 | -1 | sweetn | 700.5312 | 2 |
| 2773 | soma | 10110812 | 44656714 | 657952.0 | 265904.00 | 156680 | -1 | marquism | -6115.3125 | 1 |
| 2773 | soma | 10110812 | 44656784 | 667914.0 | 256180.00 | 145520 | -1 | marquism | 3447.3750 | 2 |
| 2773 | soma | 10234277 | 34371718 | 493445.3 | 360456.34 | 122800 | 2392 | eichlerk | -4578.6562 | 1 |
| 2773 | soma | 10234277 | 36336623 | 493429.4 | 360356.44 | 122560 | 2197 | jefferis | -4492.1562 | 2 |
| 2773 | soma | 10479161 | 35374843 | 192769.0 | 213598.00 | 202960 | 1960 | obrusnikt | -29056.5000 | 1 |
| 2773 | soma | 10479161 | 35376150 | 327465.0 | 208878.00 | 219720 | -1 | obrusnikt | 13896.2900 | 2 |
| 2773 | soma | 12173355 | 44728863 | 649485.0 | 226483.00 | 181440 | -1 | marquism | -4930.6875 | 1 |
| 2773 | soma | 12173355 | 44728872 | 648709.0 | 227851.00 | 181120 | -1 | marquism | -3965.1250 | 2 |
| 2773 | soma | 12526415 | 1112769 | 486046.6 | 126634.30 | 162360 | 1926 | adamjohn | -2019.5625 | 1 |
| 2773 | soma | 12526415 | 49202602 | 486127.0 | 127527.00 | 162600 | 3513 | gibbp | -1784.5625 | 2 |
| 2773 | soma | 12526731 | 4375796 | 600107.4 | 164816.88 | 209640 | 1787 | alis | -19557.8125 | 1 |
| 2773 | soma | 12526731 | 49752844 | 600100.0 | 164768.00 | 209640 | 2471 | gibbp | -19553.0625 | 2 |
| 2773 | soma | 12529405 | 51019292 | 432457.0 | 146688.00 | 62800 | 1948 | lia | -2623.7188 | 1 |
| 2773 | soma | 12529405 | 51021568 | 462561.0 | 275852.00 | 192680 | -1 | lia | 17362.4688 | 2 |
| 2773 | soma | 13082053 | 52561173 | 632767.4 | 284886.38 | 29440 | 2426 | toj | -13823.5625 | 1 |
| 2773 | soma | 13082053 | 52561151 | 632533.3 | 284312.50 | 30280 | -1 | toj | -13011.1875 | 2 |
| 2773 | soma | 14019704 | 44310666 | 411763.0 | 163520.42 | 79080 | 2336 | tenshawe | 979.3810 | 1 |
| 2773 | soma | 14019704 | 44310655 | 411511.6 | 163639.28 | 79320 | -1 | tenshawe | 983.9166 | 2 |
| 2773 | soma | 14462790 | 56638440 | 392006.1 | 167674.31 | 87520 | 1750 | ludwigh | 681.3281 | 1 |
| 2773 | soma | 14462790 | 57473239 | 372964.3 | 161629.42 | 106960 | -1 | ludwigh | 4346.0938 | 2 |
| 2773 | soma | 14933802 | 59013131 | 293730.8 | 147386.66 | 207680 | 2541 | kinde | -8970.6250 | 1 |
| 2773 | soma | 14933802 | 56163931 | 272602.0 | 161879.00 | 235200 | -1 | sancerg | -7160.1328 | 2 |
| 2773 | soma | 14997019 | 59362821 | 440576.4 | 190338.95 | 55000 | 3113 | kandimallap | -4747.6250 | 1 |
| 2773 | soma | 14997019 | 59844352 | 440777.4 | 183727.28 | 93760 | -1 | kandimallap | 10568.4600 | 2 |
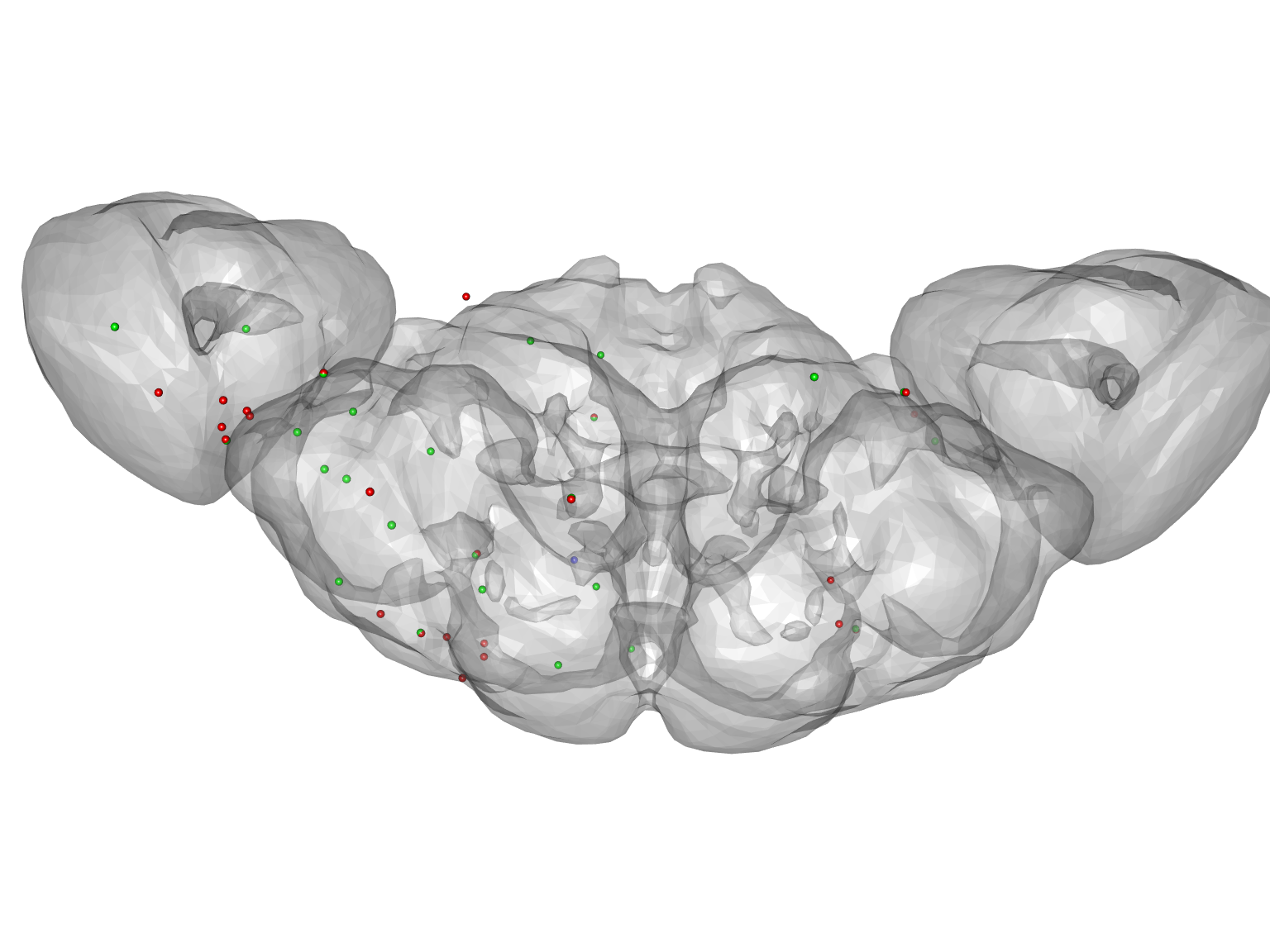
This lets us see that in some cases there are two soma tags outside the neuropil (negative d) and close together - these are probably duplicates - whereas in other cases it is likely that points were added in error. We can also plot the points colouring them by their rank order (most external first).
# make a colour palette with as many entries as the maximum number of soma # tags in a neuron pal=rainbow(max(multiple_soma_info$rank)) multiple_soma_info %>% with(expr = spheres3d(X,Y,Z, col=pal[rank], rad=2000)) plot3d(FAFB) par3d(zoom=.6)
Now we can use this information to construct an url for each node.
multiple_soma_info %>% rowwise() %>% mutate(url = open_fafb( cbind(X, Y, Z), active_skeleton_id = skeletonID, active_node_id = treenodeID, open = FALSE )) -> multiple_soma_info
It might be useful to know who ‘owns’ each neuron. I think the simplest way to assign this is by the user who has traced most nodes for each skeleton (since there may be different users responsible for each soma).
get_top_user <- function(x, ...) { ul=catmaid_get_user_list(...) # save time by checking unique skids only ddx=unique(x) gtu_one <- function(x, ...) { t <- try({ res=catmaid_get_contributor_stats(x, ...) w=which.max(res$node_contributors$n) ul$login[match(res$node_contributors$id[w], ul$id)] }) if(inherits(t, 'try-error')) NA_character_ else t } topus <- sapply(ddx, gtu_one) topus[match(x, ddx)] }
Google sheet
Let’s make a google sheet with all those urls that we can then review manually:
library(googlesheets) # helper function to upload via temp file # since writing cells via API is very slow gs_upload_tf <- function(x, ...) { tf=tempfile(fileext = '.tsv') on.exit(unlink(tf)) write.table(x, file=tf, sep="\t", row.names = FALSE) gs_upload(tf, ...) } multiple_soma_info %>% arrange(skeletonID, d) %>% group_by(skeletonID) %>% mutate(user=get_top_user(skeletonID)) %>% gs_upload_tf(sheet_title = 'multi_soma_neurons')
As an alternative we can divide that up with one worksheet per user.
library(googlesheets) gs <- googlesheets::gs_new("multi_soma_neurons_by_user") gs_add_sheet <- function(x, gs, ...) { gs_ws_new(row_extent = nrow(x)+1, col_extent = ncol(x), ss = gs, ..., input=x, col_names=T) } multiple_soma_info %>% arrange(skeletonID, d) %>% mutate(user=factor(get_top_user(skeletonID))) -> msi2 for(u in levels(msi2$user)) { gs_add_sheet(subset(msi2, user==u), gs, ws_title=u) cat(".") }