Introduction
This short article gives an example of across dataset connectivity clustering using the flywire and hemibrain datasets. This provides support for specific neuronal cell types first proposed in the hemibrain.
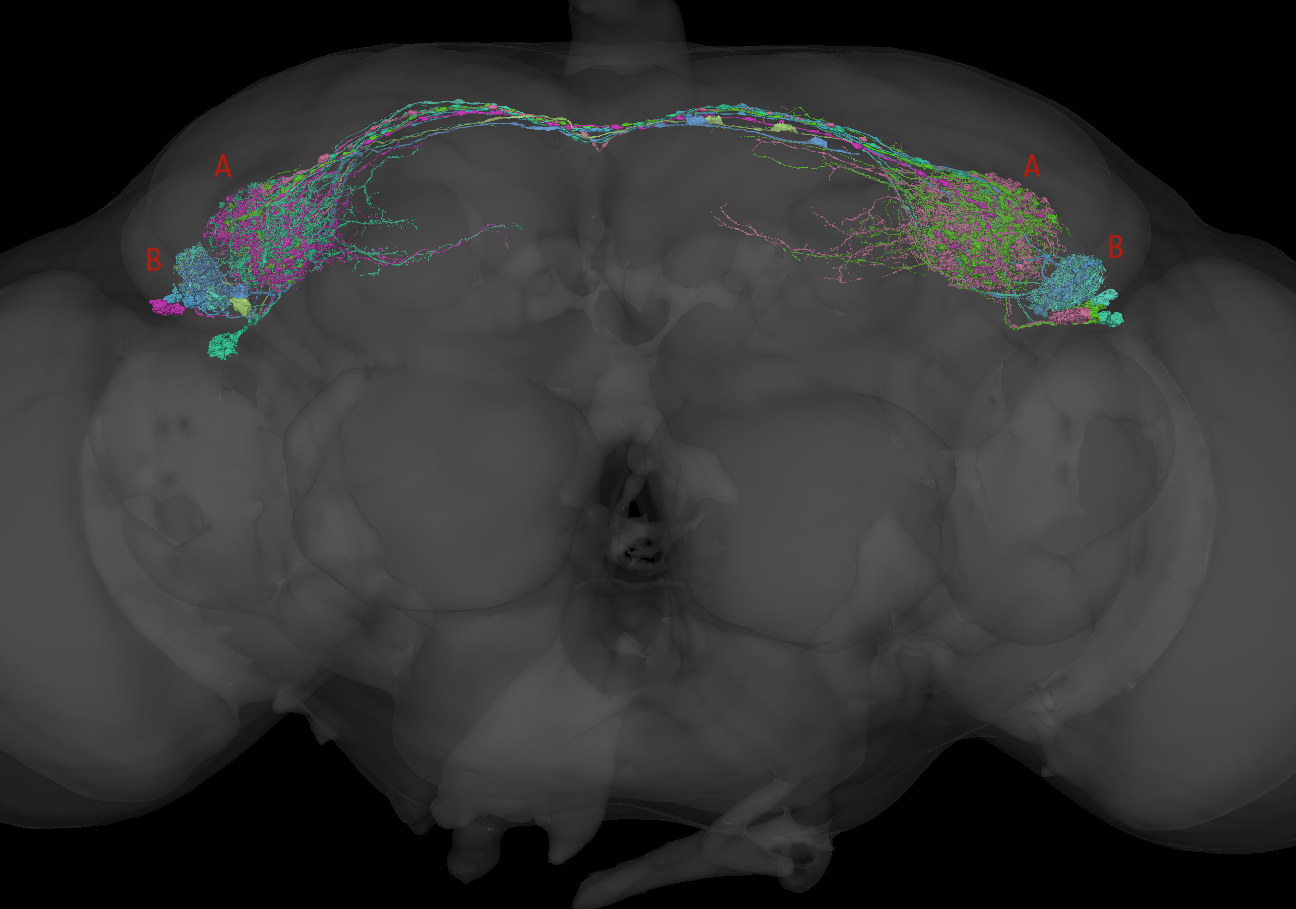
TuTu neurons are a class of visual interneurons that connect the two
optic tubercles on each side of the brain. Inputs to the tubercle
include narrow field visual interneurons e.g. those responsive to small
objects such as other flies. You can read more about them on VFB, where
they are called adult tubercle-tubercle neuron
[FBbt_00051062].
There are 4 TuTu neurons originating from each hemisphere (8 per brain). The TuTuA neurons have larger arborisations in the medial part of the optic tubercle; the TuTuB neurons have smaller arbours in the more lateral part. You can examine them in 3D using this cocoglancer scene (cocoglancer is an across-dataset configuration of the neuroglancer web app described here).
Setup
If this is your first time using coconatfly, some additional setup is required beyond package installation. Please see the getting started vignette for details of downloading / authenticating to connectome data sources.
Fetching the neurons
OK with that preamble, let’s get started. First we need to load two required libraries.
For this analysis we will use the version 630 connectivity / annotation data released in June 2023. This ensures that the cell type annotations are in a state preceding the full outcome of the analysis in this vignette. We will set an option use the lower level fafbseg package to ensure this. You may need to download the relevant data dumps if you have not done so previously (see also getting started vignette).
fafbseg::flywire_connectome_data_version(set = 630)
# fafbseg::download_flywire_release_data(version = 630)The following TuTu neurons are recorded in the hemibrain and
FAFB-FlyWire. Note the use of a regular expression (introduced by
/) to match against the type field. .* means
any number of characters can follow TuTu in the type name.
tutus=cf_meta(cf_ids('/type:TuTu.*', datasets = c("hemibrain", "flywire")))
#> Loading required namespace: git2r
tutus
#> id side class subclass subsubclass type
#> 1 720575940621226846 L central <NA> <NA> TuTuA
#> 2 720575940633550431 R central <NA> <NA> TuTuB_a
#> 3 720575940612218547 L central <NA> <NA> TuTuA
#> 4 720575940635979000 L central <NA> <NA> TuTuB_b
#> 5 720575940637997427 R central <NA> <NA> TuTuB_b
#> 6 720575940623326868 L central <NA> <NA> TuTuB_a
#> 7 720575940614041238 R central <NA> <NA> TuTuA
#> 8 720575940622538520 R central <NA> <NA> TuTuA
#> 9 5813013691 L <NA> <NA> <NA> TuTuA
#> 10 676836779 L <NA> <NA> <NA> TuTuA
#> 11 708290604 R <NA> <NA> <NA> TuTuA
#> 12 925008763 R <NA> <NA> <NA> TuTuA
#> 13 5813014028 R <NA> <NA> <NA> TuTuB_a
#> 14 770226597 L <NA> <NA> <NA> TuTuB_a
#> 15 769544445 R <NA> <NA> <NA> TuTuB_b
#> 16 987759904 L <NA> <NA> <NA> TuTuB_b
#> lineage group instance dataset key
#> 1 putative_primary <NA> TuTuA_L flywire fw:720575940621226846
#> 2 putative_primary <NA> TuTuB_a_R flywire fw:720575940633550431
#> 3 putative_primary <NA> TuTuA_L flywire fw:720575940612218547
#> 4 putative_primary <NA> TuTuB_b_L flywire fw:720575940635979000
#> 5 putative_primary <NA> TuTuB_b_R flywire fw:720575940637997427
#> 6 putative_primary <NA> TuTuB_a_L flywire fw:720575940623326868
#> 7 putative_primary <NA> TuTuA_R flywire fw:720575940614041238
#> 8 putative_primary <NA> TuTuA_R flywire fw:720575940622538520
#> 9 <NA> <NA> TuTuA(ADL19)_L hemibrain hb:5813013691
#> 10 <NA> <NA> TuTuA(ADL19)_L hemibrain hb:676836779
#> 11 ADL19 <NA> TuTuA_R hemibrain hb:708290604
#> 12 ADL19 <NA> TuTuA_R hemibrain hb:925008763
#> 13 ADL19 <NA> TuTuB_a_R hemibrain hb:5813014028
#> 14 <NA> <NA> TuTuB_a(ADL19)_L hemibrain hb:770226597
#> 15 ADL19 <NA> TuTuB_b_R hemibrain hb:769544445
#> 16 <NA> <NA> TuTuB_b(ADL19)_L hemibrain hb:987759904
tutus %>%
count(dataset, type, side)
#> dataset type side n
#> 1 flywire TuTuA L 2
#> 2 flywire TuTuA R 2
#> 3 flywire TuTuB_a L 1
#> 4 flywire TuTuB_a R 1
#> 5 flywire TuTuB_b L 1
#> 6 flywire TuTuB_b R 1
#> 7 hemibrain TuTuA L 2
#> 8 hemibrain TuTuA R 2
#> 9 hemibrain TuTuB_a L 1
#> 10 hemibrain TuTuB_a R 1
#> 11 hemibrain TuTuB_b L 1
#> 12 hemibrain TuTuB_b R 1So we can see the hemibrain TuTuB neurons have been typed into two subtypes, but the flywire neurons have not. We can run across-dataset cosine connectivity clustering on these neurons like so:
tutus %>%
cf_cosine_plot()
#> Matching types across datasets. Keeping 638/2063 output connections with total weight 13502/35096 (38%)
#> Matching types across datasets. Keeping 908/2536 input connections with total weight 18626/44807 (42%)
#> Warning in Matrix::cov2cor(cpx): diag(V) has non-positive or non-finite
#> entries; finite result is doubtful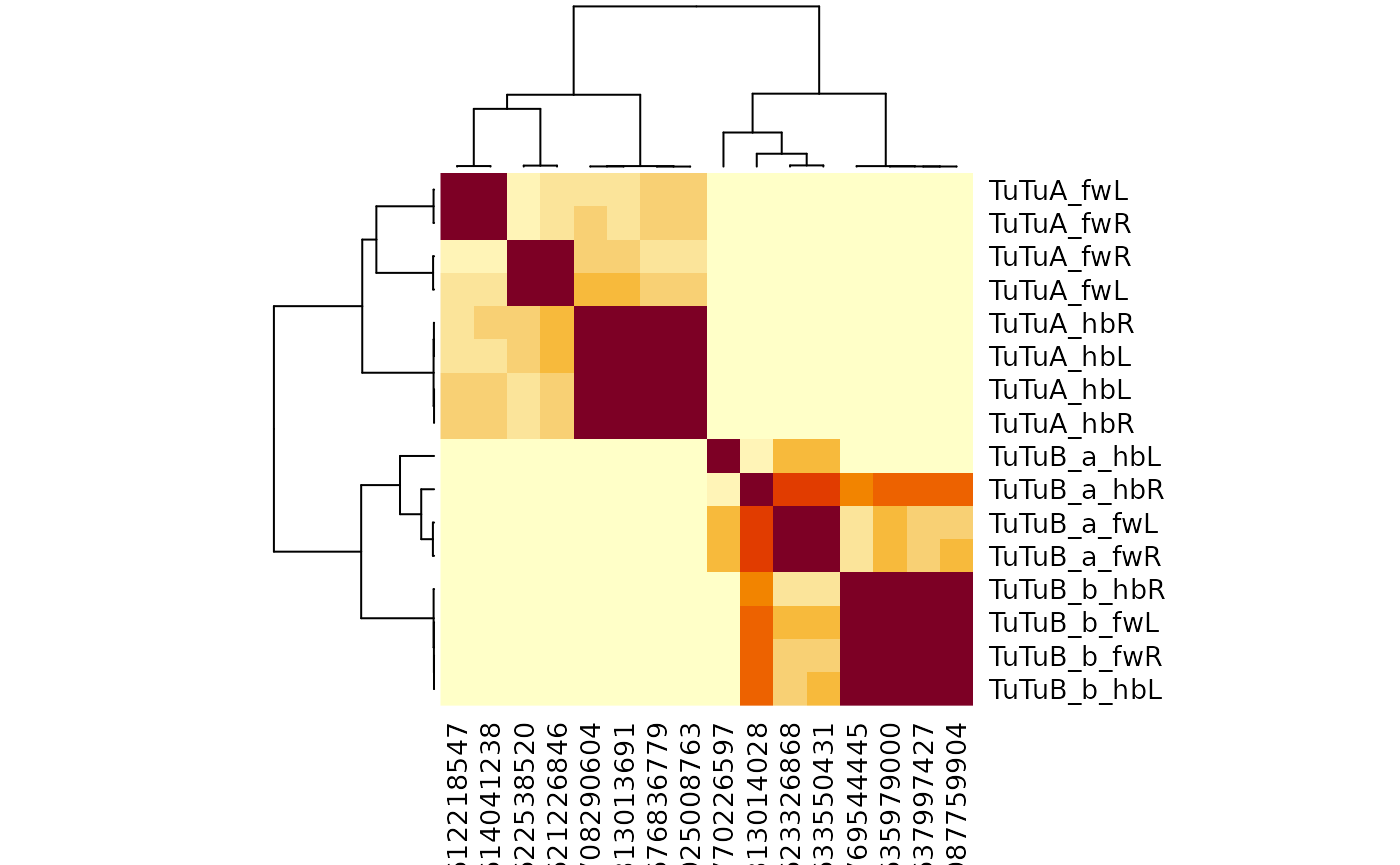
So this is interesting at a couple of levels. First we can see that
the TuTuB hemibrain neurons separate into two groups. This is true even
though things are a bit messy for the TuTuB_a_hbL neuron
which will almost certainly be heavily truncated as it has its soma on
the left hand (missing) side of the hemibrain.
We can also see the flywire neurons also split up nicely. So we have two groups of four neurons each. Each group contains one neuron from each hemisphere x dataset combination. So we can update our flywire cell typing to match the hemibrain TuTuB_a and TuTuB_b groups.
TuTuA
However, interestingly we can also see that the TuTuA group splits very strongly in flywire into two groups that are not evident in the hemibrain. To me this strongly suggests that some property of the bilateral connectivity defines this difference.
We can try seeing if we just use the input connectivity to cluster if things resolve:
tutus %>%
filter(grepl("TuTuA", type)) %>%
cf_cosine_plot(partners = 'in')
#> Matching types across datasets. Keeping 875/1843 input connections with total weight 17636/36231 (49%)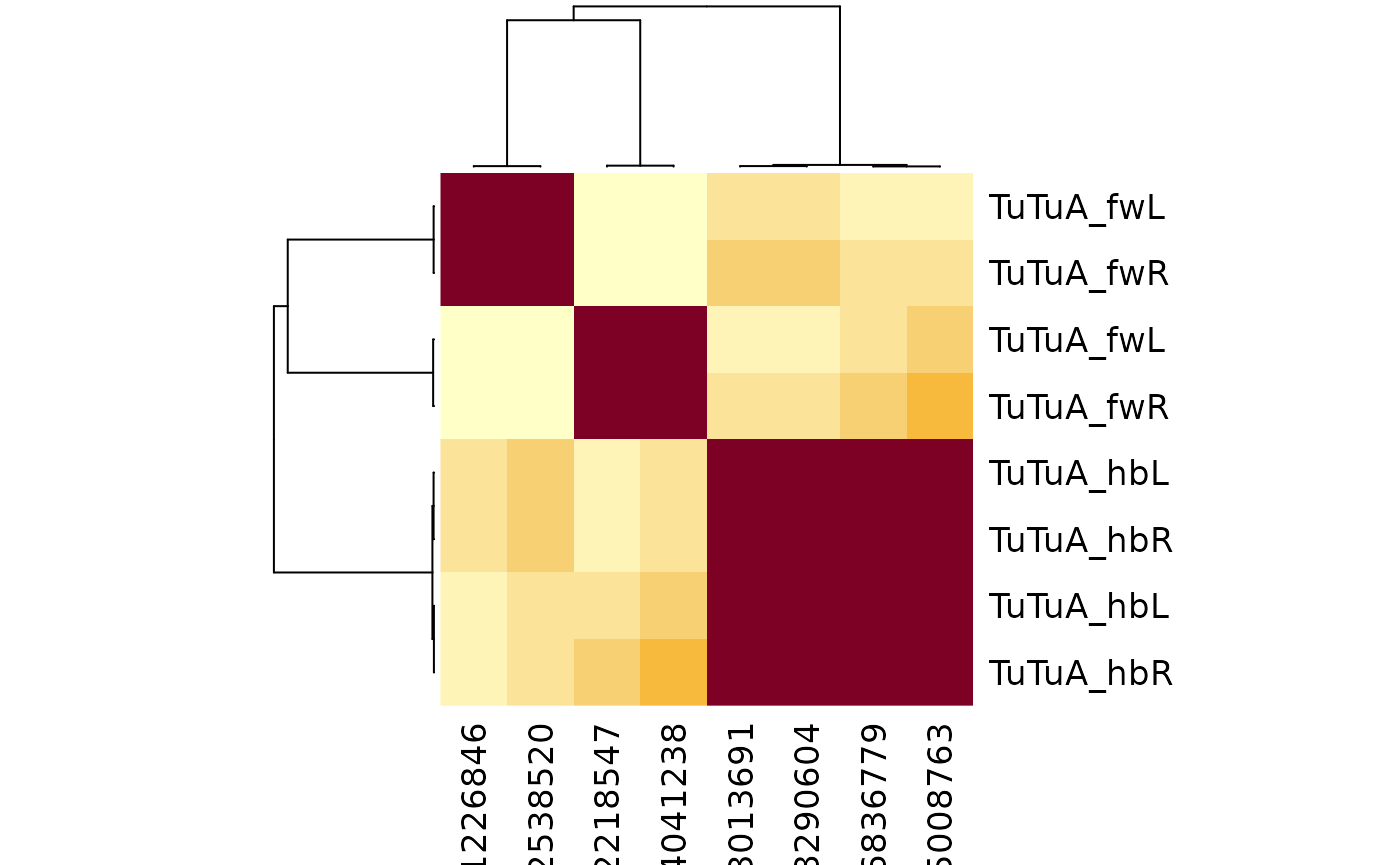
Hmm the situation is still the same. Since the two flywire groups are so well-separated this is almost certainly a real split. In fact there is a candidate anatomical difference in the placement of the contralateral axons that clearly correlates with this split.
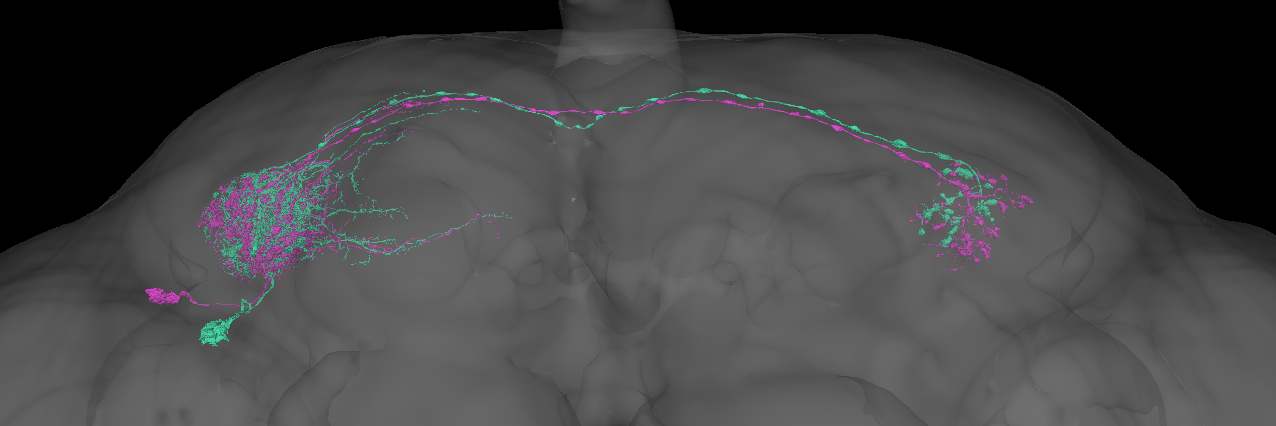
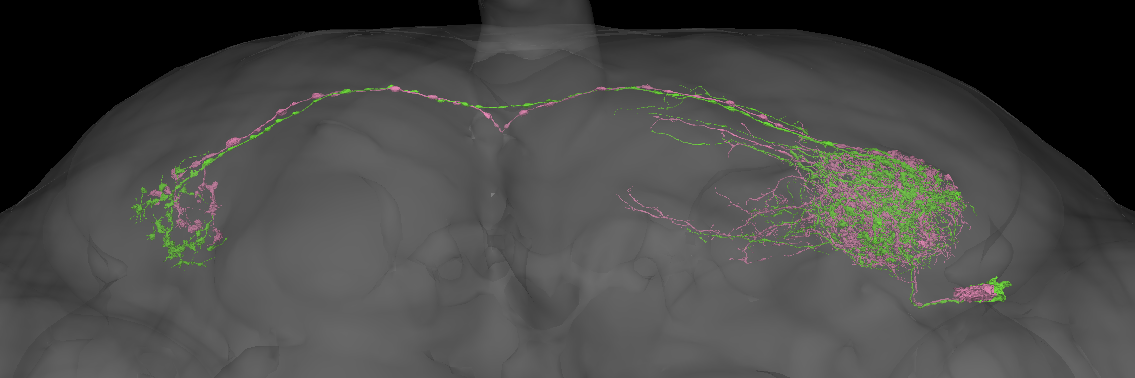
This candidate axonal difference is even visible in the hemibrain.
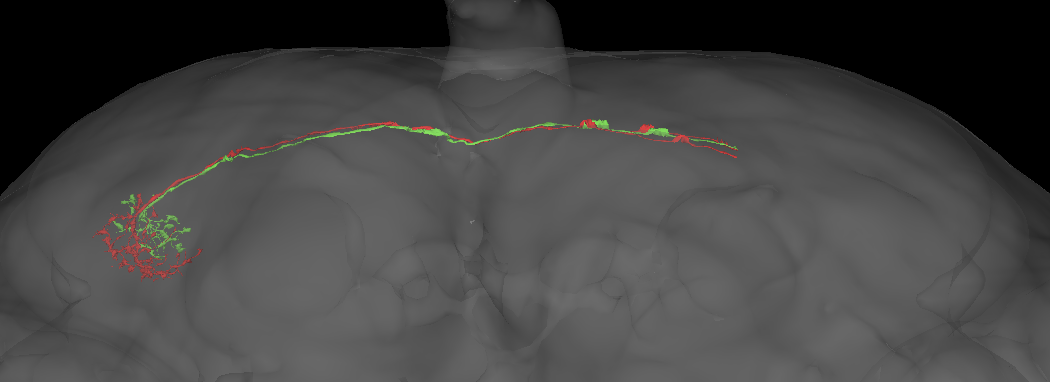
However since the across-brain connectivity clustering does not yet support this (likely owing to a lack of shared labelled partner neurons), I will not record the split for now. I strongly suspect that we will observe this split in future connectomes though.
Setting the new types
tutus.hc <- tutus %>%
cf_cosine_plot(heatmap = F)
#> Matching types across datasets. Keeping 638/2063 output connections with total weight 13502/35096 (38%)
#> Matching types across datasets. Keeping 908/2536 input connections with total weight 18626/44807 (42%)
#> Warning in Matrix::cov2cor(cpx): diag(V) has non-positive or non-finite
#> entries; finite result is doubtful
# nb as a convenience cf_meta accepts an hclust object as input
tutus.meta=cf_meta(tutus.hc)
plot(tutus.hc, labels = tutus.meta$type)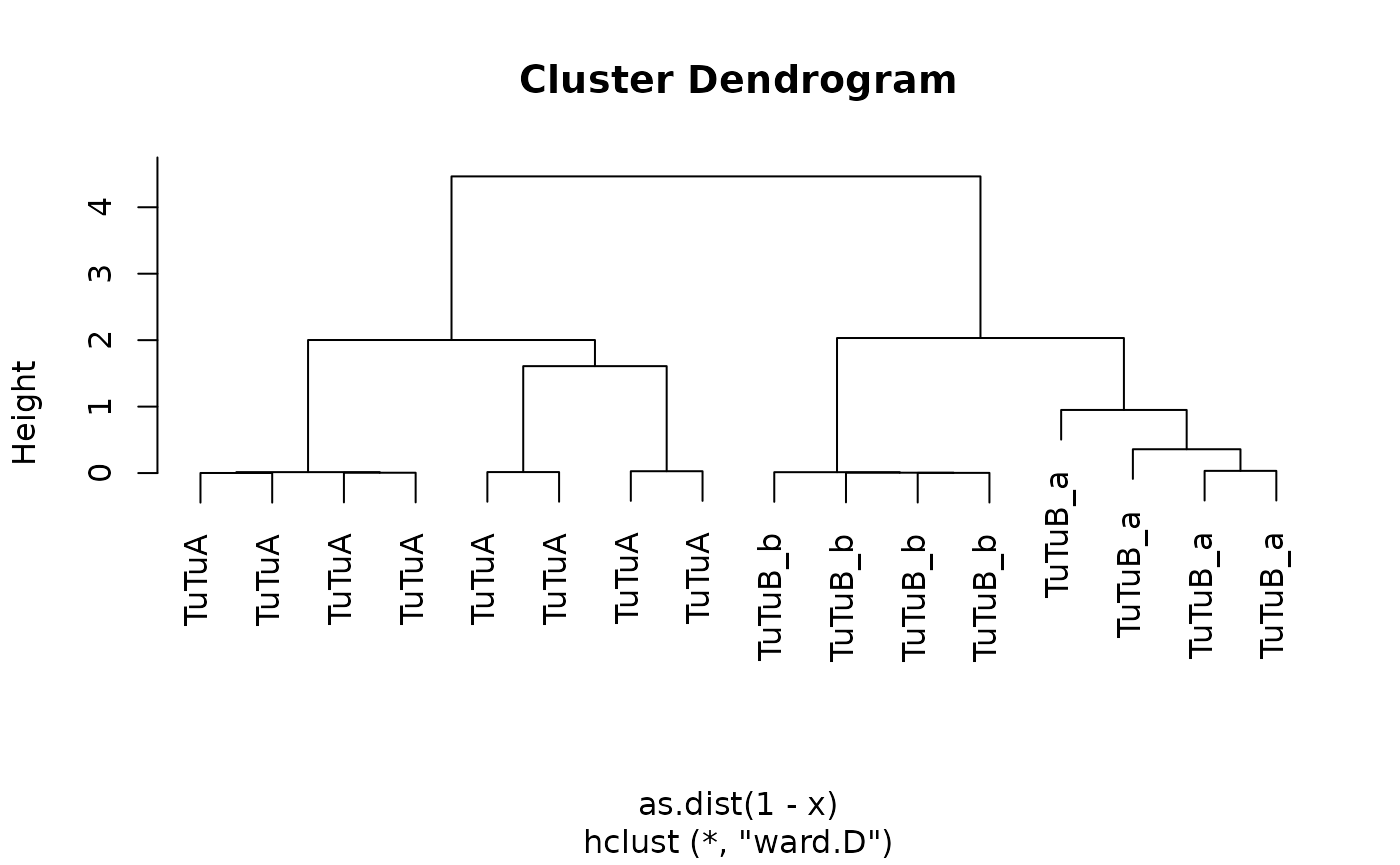
tutus.meta2=coconat::add_cluster_info(tutus.meta, tutus.hc, k = 5)
#> Warning in coconat::add_cluster_info(tutus.meta, tutus.hc, k = 5): Multiple standard id columns are present in tutus.meta
#> Choosing key
tutus.meta2 %>%
count(dataset, group_k5)
#> dataset group_k5 n
#> 1 flywire 2 2
#> 2 flywire 3 2
#> 3 flywire 4 2
#> 4 flywire 5 2
#> 5 hemibrain 1 4
#> 6 hemibrain 4 2
#> 7 hemibrain 5 2Group 4 contains the TuTuB_b neurons
tutus.meta2 %>%
filter(group_k5==4)
#> id side class subclass subsubclass type lineage
#> 1 720575940637997427 R central <NA> <NA> TuTuB_b putative_primary
#> 2 720575940635979000 L central <NA> <NA> TuTuB_b putative_primary
#> 3 769544445 R <NA> <NA> <NA> TuTuB_b ADL19
#> 4 987759904 L <NA> <NA> <NA> TuTuB_b <NA>
#> group instance dataset key dendid group_k5
#> 1 <NA> TuTuB_b_R flywire fw:720575940637997427 11 4
#> 2 <NA> TuTuB_b_L flywire fw:720575940635979000 10 4
#> 3 <NA> TuTuB_b_R hemibrain hb:769544445 9 4
#> 4 <NA> TuTuB_b(ADL19)_L hemibrain hb:987759904 12 4Group 5 contains the TuTuB_b neurons
tutus.meta2 %>%
filter(group_k5==5)
#> id side class subclass subsubclass type lineage
#> 1 720575940623326868 L central <NA> <NA> TuTuB_a putative_primary
#> 2 720575940633550431 R central <NA> <NA> TuTuB_a putative_primary
#> 3 770226597 L <NA> <NA> <NA> TuTuB_a <NA>
#> 4 5813014028 R <NA> <NA> <NA> TuTuB_a ADL19
#> group instance dataset key dendid group_k5
#> 1 <NA> TuTuB_a_L flywire fw:720575940623326868 15 5
#> 2 <NA> TuTuB_a_R flywire fw:720575940633550431 16 5
#> 3 <NA> TuTuB_a(ADL19)_L hemibrain hb:770226597 13 5
#> 4 <NA> TuTuB_a_R hemibrain hb:5813014028 14 5The following code could help find the Flybase/Virtual Fly Brain anatomy ontology (FBbt) ids for these neurons:
Old vs new annotations
In the release of the flywire annotations data prepared in December 2023 to accompany the public release of the v783 proofread FlyWire segmentation, we updated some of these TuTu cell types. This can be summarised as follows:
# nb temporarily change active flywire version
fafbseg::flywire_connectome_data_version(set = 783)
tutus.new=cf_meta(cf_ids(flywire='/type:TuTu.*'))
fafbseg::flywire_connectome_data_version(set = 630)